Prune-1 drives polarization of tumor-associated macrophages (TAMs) within the lung metastatic niche in triple-negative breast cancer
- PMID: 33426510
- PMCID: PMC7779777
- DOI: 10.1016/j.isci.2020.101938
Prune-1 drives polarization of tumor-associated macrophages (TAMs) within the lung metastatic niche in triple-negative breast cancer
Abstract
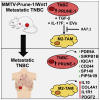
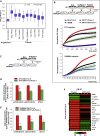
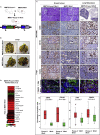
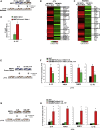
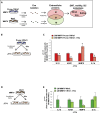
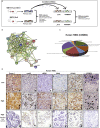
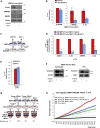
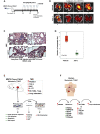
M2-tumor-associated macrophages (M2-TAMs) in the tumor microenvironment represent a prognostic indicator for poor outcome in triple-negative breast cancer (TNBC). Here we show that Prune-1 overexpression in human TNBC patients has positive correlation to lung metastasis and infiltrating M2-TAMs. Thus, we demonstrate that Prune-1 promotes lung metastasis in a genetically engineered mouse model of metastatic TNBC augmenting M2-polarization of TAMs within the tumor microenvironment. Thus, this occurs through TGF-β enhancement, IL-17F secretion, and extracellular vesicle protein content modulation. We also find murine inactivating gene variants in human TNBC patient cohorts that are involved in activation of the innate immune response, cell adhesion, apoptotic pathways, and DNA repair. Altogether, we indicate that the overexpression of Prune-1, IL-10, COL4A1, ILR1, and PDGFB, together with inactivating mutations of PDE9A, CD244, Sirpb1b, SV140, Iqca1, and PIP5K1B genes, might represent a route of metastatic lung dissemination that need future prognostic validations.
Keywords: Cancer; Immunology; Molecular Biology.
© 2020 The Author(s).
Conflict of interest statement
All the authors declare no competing financial interests.
Figures
Similar articles
-
The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer.Molecules. 2024 Mar 26;29(7):1469. doi: 10.3390/molecules29071469. Molecules. 2024. PMID: 38611749 Free PMC article.
-
CYP4A in tumor-associated macrophages promotes pre-metastatic niche formation and metastasis.Oncogene. 2017 Aug 31;36(35):5045-5057. doi: 10.1038/onc.2017.118. Epub 2017 May 8. Oncogene. 2017. PMID: 28481877 Free PMC article.
-
Triple-negative breast cancer influences a mixed M1/M2 macrophage phenotype associated with tumor aggressiveness.PLoS One. 2022 Aug 12;17(8):e0273044. doi: 10.1371/journal.pone.0273044. eCollection 2022. PLoS One. 2022. PMID: 35960749 Free PMC article.
-
Triple negative breast cancer: Key role of Tumor-Associated Macrophages in regulating the activity of anti-PD-1/PD-L1 agents.Biochim Biophys Acta Rev Cancer. 2018 Jan;1869(1):78-84. doi: 10.1016/j.bbcan.2017.10.007. Epub 2017 Nov 7. Biochim Biophys Acta Rev Cancer. 2018. PMID: 29126881 Review.
-
Subverted macrophages in the triple-negative breast cancer ecosystem.Biomed Pharmacother. 2023 Oct;166:115414. doi: 10.1016/j.biopha.2023.115414. Epub 2023 Sep 4. Biomed Pharmacother. 2023. PMID: 37660651 Review.
Cited by
-
The tumor-immune ecosystem in shaping metastasis.Am J Physiol Cell Physiol. 2023 Mar 1;324(3):C707-C717. doi: 10.1152/ajpcell.00132.2022. Epub 2023 Jan 30. Am J Physiol Cell Physiol. 2023. PMID: 36717100 Free PMC article. Review.
-
Epigenetics and immune cells in medulloblastoma.Front Genet. 2023 Mar 10;14:1135404. doi: 10.3389/fgene.2023.1135404. eCollection 2023. Front Genet. 2023. PMID: 36968588 Free PMC article. Review.
-
Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy.Cancers (Basel). 2023 Jun 16;15(12):3222. doi: 10.3390/cancers15123222. Cancers (Basel). 2023. PMID: 37370832 Free PMC article.
-
PRUNE1 and NME/NDPK family proteins influence energy metabolism and signaling in cancer metastases.Cancer Metastasis Rev. 2024 Jun;43(2):755-775. doi: 10.1007/s10555-023-10165-4. Epub 2024 Jan 5. Cancer Metastasis Rev. 2024. PMID: 38180572 Free PMC article. Review.
-
Immune Responses against Disseminated Tumor Cells.Cancers (Basel). 2021 May 21;13(11):2515. doi: 10.3390/cancers13112515. Cancers (Basel). 2021. PMID: 34063848 Free PMC article. Review.
References
-
- Boye K., Grotterod I., Aasheim H.C., Hovig E., Maelandsmo G.M. Activation of NF-kappaB by extracellular S100A4: analysis of signal transduction mechanisms and identification of target genes. Int. J. Cancer. 2008;123:1301–1310. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

