Construction of an mRNA-miRNA-lncRNA network prognostic for triple-negative breast cancer
- PMID: 33428596
- PMCID: PMC7835059
- DOI: 10.18632/aging.202254
Construction of an mRNA-miRNA-lncRNA network prognostic for triple-negative breast cancer
Abstract
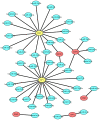
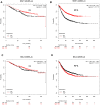
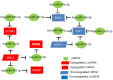
The aim of this study was to establish a novel competing endogenous RNA (ceRNA) network able to predict prognosis in patients with triple-negative breast cancer (TNBC). Differential gene expression analysis was performed using the GEO2R tool. Enrichr and STRING were used to conduct protein-protein interaction and pathway enrichment analyses, respectively. Upstream lncRNAs and miRNAs were identified using miRNet and mirTarBase, respectively. Prognostic values, expression, and correlational relationships of mRNAs, lncRNAs, and miRNAs were examined using GEPIA, starBase, and Kaplan-Meier plotter. It total, 860 upregulated and 622 downregulated differentially expressed mRNAs were identified in TNBC. Ten overexpressed and two underexpressed hub genes were screened. Next, 10 key miRNAs upstream of these key hub genes were predicted, of which six upregulated miRNAs were significantly associated with poor prognosis and four downregulated miRNAs were associated with good prognosis in TNBC. NEAT1 and MAL2 were selected as key lncRNAs. An mRNA-miRNA-lncRNA network in TNBC was constructed. Thus, we successfully established a novel mRNA-miRNA-lncRNA regulatory network, each component of which is prognostic for TNBC.
Keywords: biomarker; competing endogenous RNA; prognosis; triple-negative breast cancer.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

