Full length genomic sanger sequencing and phylogenetic analysis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Nigeria
- PMID: 33428634
- PMCID: PMC7799769
- DOI: 10.1371/journal.pone.0243271
Full length genomic sanger sequencing and phylogenetic analysis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Nigeria
Abstract
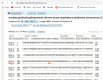
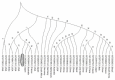
In an outbreak, effective detection of the aetiological agent(s) involved using molecular techniques is key to efficient diagnosis, early prevention and management of the spread. However, sequencing is necessary for mutation monitoring and tracking of clusters of transmission, development of diagnostics and for vaccines and drug development. Many sequencing methods are fast evolving to reduce test turn-around-time and to increase through-put compared to Sanger sequencing method; however, Sanger sequencing remains the gold standard for clinical research sequencing with its 99.99% accuracy This study sought to generate sequence data of SARS-CoV-2 using Sanger sequencing method and to characterize them for possible site(s) of mutations. About 30 pairs of primers were designed, synthesized, and optimized using endpoint PCR to generate amplicons for the full length of the virus. Cycle sequencing using BigDye Terminator v.3.1 and capillary gel electrophoresis on ABI 3130xl genetic analyser were performed according to the manufacturers' instructions. The sequence data generated were assembled and analysed for variations using DNASTAR Lasergene 17 SeqMan Ultra. Total length of 29,760bp of SARS-CoV-2 was assembled from the sample analysed and deposited in GenBank with accession number: MT576584. Blast result of the sequence assembly shows a 99.97% identity with the reference sequence. Variations were noticed at positions: nt201, nt2997, nt14368, nt16535, nt20334, and nt28841-28843, which caused amino acid alterations at the S (aa614) and N (aa203-204) regions. The mutations observed at S and N-gene in this study may be indicative of a gradual changes in the genetic coding of the virus hence, the need for active surveillance of the viral genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

