PfAgo-based detection of SARS-CoV-2
- PMID: 33429204
- PMCID: PMC7832551
- DOI: 10.1016/j.bios.2020.112932
PfAgo-based detection of SARS-CoV-2
Abstract
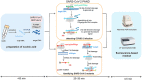
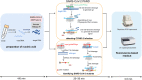
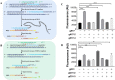
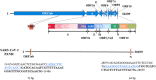
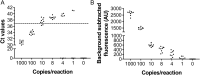
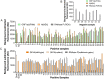
In the present study, we upgraded Pyrococcus furiosus Argonaute (PfAgo) mediated nucleic acid detection method and established a highly sensitive and accurate molecular diagnosis platform for the large-scale screening of COVID-19 infection. Briefly, RT-PCR was performed with the viral RNA extracted from nasopharyngeal or oropharyngeal swabs as template to amplify conserved regions in the viral genome. Next, PfAgo, guide DNAs and molecular beacons in appropriate buffer were added to the PCR products, followed by incubating at 95 °C for 20-30 min. Subsequently, the fluorescence signal was detected. This method was named as SARS-CoV-2 PAND. The whole procedure is accomplished in approximately an hour with the using time of the Real-time fluorescence quantitative PCR instrument shortened from >1 h to only 3-5 min per batch in comparison with RT-qPCR, hence the shortage of the expensive Real-time PCR instrument is alleviated. Moreover, this platform was also applied to identify SARS-CoV-2 D614G mutant due to its single-nucleotide specificity. The diagnostic results of clinic samples with SARS-CoV-2 PAND displayed 100% consistence with RT-qPCR test.
Keywords: Molecular diagnosis; PfAgo; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
A Fast and Sensitive One-Tube SARS-CoV-2 Detection Platform Based on RTX-PCR and Pyrococcus furiosus Argonaute.Biosensors (Basel). 2024 May 13;14(5):245. doi: 10.3390/bios14050245. Biosensors (Basel). 2024. PMID: 38785719 Free PMC article.
-
Rapid detection of SARS-CoV-2 variants by molecular-clamping technology-based RT-qPCR.Microbiol Spectr. 2024 Nov 5;12(11):e0424823. doi: 10.1128/spectrum.04248-23. Epub 2024 Oct 16. Microbiol Spectr. 2024. PMID: 39412285 Free PMC article.
-
Diagnosis with Metagenomic Next-Generation Sequencing (mNGS) technology and real-time PCR for SARS-CoV-2 Omicron detection using various nasopharyngeal swabs in SARS-CoV-2 Omicron.PLoS One. 2024 Aug 6;19(8):e0305289. doi: 10.1371/journal.pone.0305289. eCollection 2024. PLoS One. 2024. PMID: 39106263 Free PMC article.
-
Molecular Diagnostic Tools for the Detection of SARS-CoV-2.Int Rev Immunol. 2021;40(1-2):143-156. doi: 10.1080/08830185.2020.1871477. Epub 2021 Jan 13. Int Rev Immunol. 2021. PMID: 33439059 Review.
-
Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis.J Med Virol. 2021 Feb;93(2):719-725. doi: 10.1002/jmv.26349. Epub 2020 Aug 2. J Med Virol. 2021. PMID: 32706393 Free PMC article.
Cited by
-
ANCA: artificial nucleic acid circuit with argonaute protein for one-step isothermal detection of antibiotic-resistant bacteria.Nat Commun. 2023 Dec 5;14(1):8033. doi: 10.1038/s41467-023-43899-4. Nat Commun. 2023. PMID: 38052830 Free PMC article.
-
Pyrococcus furiosus Argonaute-mediated porcine epidemic diarrhea virus detection.Appl Microbiol Biotechnol. 2024 Dec;108(1):137. doi: 10.1007/s00253-023-12919-0. Epub 2024 Jan 15. Appl Microbiol Biotechnol. 2024. PMID: 38229331 Free PMC article.
-
Rapid and sensitive detection of methicillin-resistant Staphylococcus aureus through the RPA-PfAgo system.Front Microbiol. 2024 Aug 21;15:1422574. doi: 10.3389/fmicb.2024.1422574. eCollection 2024. Front Microbiol. 2024. PMID: 39234537 Free PMC article.
-
Using Recombinase-Aid Amplification Combined with Pyrococcus furiosus Argonaute for Rapid Sex Identification in Flamingo (Phoenicopteridae).Animals (Basel). 2024 Dec 24;15(1):7. doi: 10.3390/ani15010007. Animals (Basel). 2024. PMID: 39794950 Free PMC article.
-
A rapid nucleic acid detection platform based on phosphorothioate-DNA and sulfur binding domain.Synth Syst Biotechnol. 2023 Feb 11;8(2):213-219. doi: 10.1016/j.synbio.2023.02.002. eCollection 2023 Jun. Synth Syst Biotechnol. 2023. PMID: 36875498 Free PMC article.
References
-
- Chen G.-Q., Choi I., Ramachandran B., Gouaux J.E. J. Am. Chem. Soc. 1994;116:8799–8800.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

