DeepCNV: a deep learning approach for authenticating copy number variations
- PMID: 33429424
- PMCID: PMC8681111
- DOI: 10.1093/bib/bbaa381
DeepCNV: a deep learning approach for authenticating copy number variations
Abstract
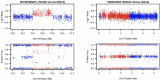
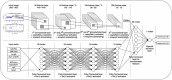
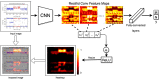
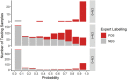
Copy number variations (CNVs) are an important class of variations contributing to the pathogenesis of many disease phenotypes. Detecting CNVs from genomic data remains difficult, and the most currently applied methods suffer from an unacceptably high false positive rate. A common practice is to have human experts manually review original CNV calls for filtering false positives before further downstream analysis or experimental validation. Here, we propose DeepCNV, a deep learning-based tool, intended to replace human experts when validating CNV calls, focusing on the calls made by one of the most accurate CNV callers, PennCNV. The sophistication of the deep neural network algorithm is enriched with over 10 000 expert-scored samples that are split into training and testing sets. Variant confidence, especially for CNVs, is a main roadblock impeding the progress of linking CNVs with the disease. We show that DeepCNV adds to the confidence of the CNV calls with an optimal area under the receiver operating characteristic curve of 0.909, exceeding other machine learning methods. The superiority of DeepCNV was also benchmarked and confirmed using an experimental wet-lab validation dataset. We conclude that the improvement obtained by DeepCNV results in significantly fewer false positive results and failures to replicate the CNV association results.
Keywords: copy number variation; deep learning.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

