Utilizing Amino Acid Composition and Entropy of Potential Open Reading Frames to Identify Protein-Coding Genes
- PMID: 33429904
- PMCID: PMC7827183
- DOI: 10.3390/microorganisms9010129
Utilizing Amino Acid Composition and Entropy of Potential Open Reading Frames to Identify Protein-Coding Genes
Abstract
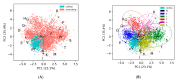
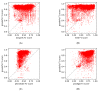
One of the main steps in gene-finding in prokaryotes is determining which open reading frames encode for a protein, and which occur by chance alone. There are many different methods to differentiate the two; the most prevalent approach is using shared homology with a database of known genes. This method presents many pitfalls, most notably the catch that you only find genes that you have seen before. The four most popular prokaryotic gene-prediction programs (GeneMark, Glimmer, Prodigal, Phanotate) all use a protein-coding training model to predict protein-coding genes, with the latter three allowing for the training model to be created ab initio from the input genome. Different methods are available for creating the training model, and to increase the accuracy of such tools, we present here GOODORFS, a method for identifying protein-coding genes within a set of all possible open reading frames (ORFS). Our workflow begins with taking the amino acid frequencies of each ORF, calculating an entropy density profile (EDP), using KMeans to cluster the EDPs, and then selecting the cluster with the lowest variation as the coding ORFs. To test the efficacy of our method, we ran GOODORFS on 14,179 annotated phage genomes, and compared our results to the initial training-set creation step of four other similar methods (Glimmer, MED2, PHANOTATE, Prodigal). We found that GOODORFS was the most accurate (0.94) and had the best F1-score (0.85), while Glimmer had the highest precision (0.92) and PHANOTATE had the highest recall (0.96).
Keywords: annotation; clustering; gene; genome; machine learning; phage; prediction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Jou W.M., Molemans F., Raeymaekers A., Berghe A.V.D., et al. Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene. Nat. Cell Biol. 1976;260:500–507. doi: 10.1038/260500a0. - DOI - PubMed
-
- Borodovsky M., McIninch J. GENMARK: Parallel gene recognition for both DNA strands. Comput. Chem. 1993;17:123–133. doi: 10.1016/0097-8485(93)85004-V. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

