Proteomic and Functional Studies Reveal Detyrosinated Tubulin as Treatment Target in Sarcomere Mutation-Induced Hypertrophic Cardiomyopathy
- PMID: 33430602
- PMCID: PMC7819533
- DOI: 10.1161/CIRCHEARTFAILURE.120.007022
Proteomic and Functional Studies Reveal Detyrosinated Tubulin as Treatment Target in Sarcomere Mutation-Induced Hypertrophic Cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is the most common genetic heart disease. While ≈50% of patients with HCM carry a sarcomere gene mutation (sarcomere mutation-positive, HCMSMP), the genetic background is unknown in the other half of the patients (sarcomere mutation-negative, HCMSMN). Genotype-specific differences have been reported in cardiac function. Moreover, HCMSMN patients have later disease onset and a better prognosis than HCMSMP patients. To define if genotype-specific derailments at the protein level may explain the heterogeneity in disease development, we performed a proteomic analysis in cardiac tissue from a clinically well-phenotyped HCM patient group.
Methods: A proteomics screen was performed in cardiac tissue from 39 HCMSMP patients, 11HCMSMN patients, and 8 nonfailing controls. Patients with HCM had obstructive cardiomyopathy with left ventricular outflow tract obstruction and diastolic dysfunction. A novel MYBPC32373insG mouse model was used to confirm functional relevance of our proteomic findings.
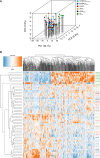
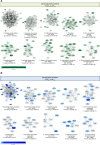
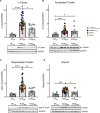
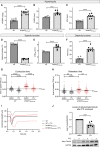
Results: In all HCM patient samples, we found lower levels of metabolic pathway proteins and higher levels of extracellular matrix proteins. Levels of total and detyrosinated α-tubulin were markedly higher in HCMSMP than in HCMSMN and controls. Higher tubulin detyrosination was also found in 2 unrelated MYBPC3 mouse models and its inhibition with parthenolide normalized contraction and relaxation time of isolated cardiomyocytes.
Conclusions: Our findings indicate that microtubules and especially its detyrosination contribute to the pathomechanism of patients with HCMSMP. This is of clinical importance since it represents a potential treatment target to improve cardiac function in patients with HCMSMP, whereas a beneficial effect may be limited in patients with HCMSMN.
Keywords: cardiomyopathies; genotype; heart diseases; mutation; proteomics; treatment; tubulin.
Conflict of interest statement
None.
Figures


Comment in
-
Tubulin Detyrosination: An Emerging Therapeutic Target in Hypertrophic Cardiomyopathy.Circ Heart Fail. 2021 Jan;14(1):e008006. doi: 10.1161/CIRCHEARTFAILURE.120.008006. Epub 2021 Jan 12. Circ Heart Fail. 2021. PMID: 33430601 Free PMC article. No abstract available.
References
-
- Ingles J, Burns C, Barratt A, Semsarian C. Application of genetic testing in hypertrophic cardiomyopathy for preclinical disease detection. Circ Cardiovasc Genet. 2015;8:852–859. doi: 10.1161/CIRCGENETICS.115.001093 - PubMed
-
- Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, et al. ; Authors/Task Force. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35:2733–2779. doi: 10.1093/eurheartj/ehu284 - PubMed
-
- Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, et al. ; EUROGENE Heart Failure Project. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54 - PubMed
-
- Vakrou S, Fukunaga R, Foster DB, Sorensen L, Liu Y, Guan Y, Woldemichael K, Pineda-Reyes R, Liu T, Tardiff JC, et al. Allele-specific differences in transcriptome, miRNome, and mitochondrial function in two hypertrophic cardiomyopathy mouse models. JCI Insight. 2018;3doi: 10.1172/jci.insight.94493 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

