A deep learning model for diagnosing dystrophinopathies on thigh muscle MRI images
- PMID: 33430797
- PMCID: PMC7798322
- DOI: 10.1186/s12883-020-02036-0
A deep learning model for diagnosing dystrophinopathies on thigh muscle MRI images
Abstract
Background: Dystrophinopathies are the most common type of inherited muscular diseases. Muscle biopsy and genetic tests are effective to diagnose the disease but cost much more than primary hospitals can reach. The more available muscle MRI is promising but its diagnostic results highly depends on doctors' experiences. This study intends to explore a way of deploying a deep learning model for muscle MRI images to diagnose dystrophinopathies.
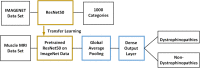
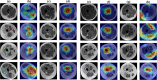
Methods: This study collected 2536 T1WI images from 432 cases who had been diagnosed by genetic analysis and/or muscle biopsy, including 148 cases with dystrophinopathies and 284 cases with other diseases. The data was randomly divided into three sets: the data from 233 cases were used to train the CNN model, the data from 97 cases for the validation experiments, and the data from 102 cases for the test experiments. We also validated our models expertise at diagnosing by comparing the model's results on the 102 cases with those of three skilled radiologists.
Results: The proposed model achieved 91% (95% CI: 0.88, 0.93) accuracy on the test set, higher than the best accuracy of 84% in radiologists. It also performed better than the skilled radiologists in sensitivity : sensitivities of the models and the doctors were 0.89 (95% CI: 0.85 0.93) versus 0.79 (95% CI:0.73, 0.84; p = 0.190).
Conclusions: The deep model achieved excellent accuracy and sensitivity in identifying cases with dystrophinopathies. The comparable performance of the model and skilled radiologists demonstrates the potential application of the model in diagnosing dystrophinopathies through MRI images.
Keywords: Computer-Assisted Diagnosis; Deep Learning; Magnetic Resonance Imaging; Muscular Diseases.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet.PLoS Med. 2018 Nov 27;15(11):e1002699. doi: 10.1371/journal.pmed.1002699. eCollection 2018 Nov. PLoS Med. 2018. PMID: 30481176 Free PMC article.
-
The trefoil with single fruit sign in muscle magnetic resonance imaging is highly specific for dystrophinopathies.Eur J Radiol. 2015 Oct;84(10):1992-8. doi: 10.1016/j.ejrad.2015.06.011. Epub 2015 Jun 18. Eur J Radiol. 2015. PMID: 26119801
-
Value of muscle magnetic resonance imaging in the differential diagnosis of muscular dystrophies related to the dystrophin-glycoprotein complex.Orphanet J Rare Dis. 2019 Nov 12;14(1):250. doi: 10.1186/s13023-019-1242-y. Orphanet J Rare Dis. 2019. PMID: 31747956 Free PMC article.
-
Accuracy of a machine learning muscle MRI-based tool for the diagnosis of muscular dystrophies.Neurology. 2020 Mar 10;94(10):e1094-e1102. doi: 10.1212/WNL.0000000000009068. Epub 2020 Feb 6. Neurology. 2020. PMID: 32029545
-
Two decades of advances in muscle imaging in children: from pattern recognition of muscle diseases to quantification and machine learning approaches.Neuromuscul Disord. 2021 Oct;31(10):1038-1050. doi: 10.1016/j.nmd.2021.08.006. Epub 2021 Oct 9. Neuromuscul Disord. 2021. PMID: 34736625 Review.
Cited by
-
Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model.Bioengineering (Basel). 2024 Jun 7;11(6):580. doi: 10.3390/bioengineering11060580. Bioengineering (Basel). 2024. PMID: 38927816 Free PMC article.
-
Advanced Deep Learning and Machine Learning Techniques for MRI Brain Tumor Analysis: A Review.Sensors (Basel). 2025 Apr 26;25(9):2746. doi: 10.3390/s25092746. Sensors (Basel). 2025. PMID: 40363185 Free PMC article. Review.
-
SEMPAI: a Self-Enhancing Multi-Photon Artificial Intelligence for Prior-Informed Assessment of Muscle Function and Pathology.Adv Sci (Weinh). 2023 Oct;10(28):e2206319. doi: 10.1002/advs.202206319. Epub 2023 Aug 15. Adv Sci (Weinh). 2023. PMID: 37582656 Free PMC article.
-
Roles of Skeletal Muscle in Development: A Bioinformatics and Systems Biology Overview.Adv Anat Embryol Cell Biol. 2023;236:21-55. doi: 10.1007/978-3-031-38215-4_2. Adv Anat Embryol Cell Biol. 2023. PMID: 37955770
-
Machine learning-based radiomics using MRI to differentiate early-stage Duchenne and Becker muscular dystrophy in children.BMC Musculoskelet Disord. 2025 Mar 22;26(1):287. doi: 10.1186/s12891-025-08538-7. BMC Musculoskelet Disord. 2025. PMID: 40121488 Free PMC article.
References
-
- Bushby K, Finkel R, Birnkrant DJ, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management[J] Lancet Neurol. 2010;9(1):77–93. - PubMed
-
- Voit T, Stuettgen P, Cremer M, et al. Dystrophin as a diagnostic marker in Duchenne and Becker muscular dystrophy. Correlation of immunofluorescence and western blot[J] Neuropediatrics. 1991;22(03):152–62. - PubMed
-
- Liu G, Jong Y, Chiang C, et al. Duchenne muscular dystrophy: MR grading system with functional correlation[J] Radiology. 1993;186(2):475–80. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

