Ventilator-associated pneumonia in critically ill patients with COVID-19
- PMID: 33430915
- PMCID: PMC7797892
- DOI: 10.1186/s13054-021-03460-5
Ventilator-associated pneumonia in critically ill patients with COVID-19
Erratum in
-
Correction to: Ventilator-associated pneumonia in critically ill patients with COVID-19.Crit Care. 2021 Apr 6;25(1):130. doi: 10.1186/s13054-021-03560-2. Crit Care. 2021. PMID: 33823901 Free PMC article. No abstract available.
Abstract
Background: Pandemic COVID-19 caused by the coronavirus SARS-CoV-2 has a high incidence of patients with severe acute respiratory syndrome (SARS). Many of these patients require admission to an intensive care unit (ICU) for invasive ventilation and are at significant risk of developing a secondary, ventilator-associated pneumonia (VAP).
Objectives: To study the incidence of VAP and bacterial lung microbiome composition of ventilated COVID-19 and non-COVID-19 patients.
Methods: In this retrospective observational study, we compared the incidence of VAP and secondary infections using a combination of microbial culture and a TaqMan multi-pathogen array. In addition, we determined the lung microbiome composition using 16S RNA analysis in a subset of samples. The study involved 81 COVID-19 and 144 non-COVID-19 patients receiving invasive ventilation in a single University teaching hospital between March 15th 2020 and August 30th 2020.
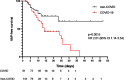
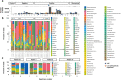
Results: COVID-19 patients were significantly more likely to develop VAP than patients without COVID (Cox proportional hazard ratio 2.01 95% CI 1.14-3.54, p = 0.0015) with an incidence density of 28/1000 ventilator days versus 13/1000 for patients without COVID (p = 0.009). Although the distribution of organisms causing VAP was similar between the two groups, and the pulmonary microbiome was similar, we identified 3 cases of invasive aspergillosis amongst the patients with COVID-19 but none in the non-COVID-19 cohort. Herpesvirade activation was also numerically more frequent amongst patients with COVID-19.
Conclusion: COVID-19 is associated with an increased risk of VAP, which is not fully explained by the prolonged duration of ventilation. The pulmonary dysbiosis caused by COVID-19, and the causative organisms of secondary pneumonia observed are similar to that seen in critically ill patients ventilated for other reasons.
Keywords: COVID-19; Critical care; Molecular diagnostics; Nosocomial infections; SARS-CoV-2; Ventilator-associated pneumonia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

