The chemfp project
- PMID: 33430977
- PMCID: PMC6896769
- DOI: 10.1186/s13321-019-0398-8
The chemfp project
Abstract
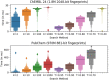
The chemfp project has had four main goals: (1) promote the FPS format as a text-based exchange format for dense binary cheminformatics fingerprints, (2) develop a high-performance implementation of the BitBound algorithm that could be used as an effective baseline to benchmark new similarity search implementations, (3) experiment with funding a pure open source software project through commercial sales, and (4) publish the results and lessons learned as a guide for future implementors. The FPS format has had only minor success, though it did influence development of the FPB binary format, which is faster to load but more complex. Both are summarized. The chemfp benchmark and the no-cost/open source version of chemfp are proposed as a reference baseline to evaluate the effectiveness of other similarity search tools. They are used to evaluate the faster commercial version of chemfp, which can test 130 million 1024-bit fingerprint Tanimotos per second on a single core of a standard x86-64 server machine. When combined with the BitBound algorithm, a k = 1000 nearest-neighbor search of the 1.8 million 2048-bit Morgan fingerprints of ChEMBL 24 averages 27 ms/query. The same search of 970 million PubChem fingerprints averages 220 ms/query, making chemfp one of the fastest CPU-based similarity search implementations. Modern CPUs are fast enough that memory bandwidth and latency are now important factors. Single-threaded search uses most of the available memory bandwidth. Sorting the fingerprints by popcount improves memory coherency, which when combined with 4 OpenMP threads makes it possible to construct an N × N similarity matrix for 1 million fingerprints in about 30 min. These observations may affect the interpretation of previous publications which assumed that search was strongly CPU bound. The chemfp project funding came from selling a purely open-source software product. Several product business models were tried, but none proved sustainable. Some of the experiences are discussed, in order to contribute to the ongoing conversation on the role of open source software in cheminformatics.
Keywords: FOSS; Format; High-performance; Molecular fingerprints; Open source; Performance benchmark; Similarity searching; Tanimoto.
Conflict of interest statement
The author declares no competing interests.
Figures


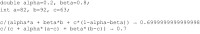
Similar articles
-
jCompoundMapper: An open source Java library and command-line tool for chemical fingerprints.J Cheminform. 2011 Jan 10;3(1):3. doi: 10.1186/1758-2946-3-3. J Cheminform. 2011. PMID: 21219648 Free PMC article.
-
Comparing structural fingerprints using a literature-based similarity benchmark.J Cheminform. 2016 Jul 5;8:36. doi: 10.1186/s13321-016-0148-0. eCollection 2016. J Cheminform. 2016. PMID: 27382417 Free PMC article.
-
Ligand-based target prediction with signature fingerprints.J Chem Inf Model. 2014 Oct 27;54(10):2647-53. doi: 10.1021/ci500361u. Epub 2014 Oct 3. J Chem Inf Model. 2014. PMID: 25230336
-
Fingerprint design and engineering strategies: rationalizing and improving similarity search performance.Future Med Chem. 2012 Oct;4(15):1945-59. doi: 10.4155/fmc.12.126. Future Med Chem. 2012. PMID: 23088275 Review.
-
Molecular fingerprint similarity search in virtual screening.Methods. 2015 Jan;71:58-63. doi: 10.1016/j.ymeth.2014.08.005. Epub 2014 Aug 15. Methods. 2015. PMID: 25132639 Review.
Cited by
-
Automated Exploration of Prebiotic Chemical Reaction Space: Progress and Perspectives.Life (Basel). 2021 Oct 26;11(11):1140. doi: 10.3390/life11111140. Life (Basel). 2021. PMID: 34833016 Free PMC article. Review.
-
BonMOLière: Small-Sized Libraries of Readily Purchasable Compounds, Optimized to Produce Genuine Hits in Biological Screens across the Protein Space.Int J Mol Sci. 2021 Jul 21;22(15):7773. doi: 10.3390/ijms22157773. Int J Mol Sci. 2021. PMID: 34360558 Free PMC article.
-
DrugGym: A testbed for the economics of autonomous drug discovery.bioRxiv [Preprint]. 2024 Jun 2:2024.05.28.596296. doi: 10.1101/2024.05.28.596296. bioRxiv. 2024. PMID: 38854082 Free PMC article. Preprint.
-
The application of machine learning methods to the prediction of novel ligands for RORγ/RORγT receptors.Comput Struct Biotechnol J. 2023 Oct 29;21:5491-5505. doi: 10.1016/j.csbj.2023.10.021. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38022699 Free PMC article.
-
Machine Learning-Boosted Docking Enables the Efficient Structure-Based Virtual Screening of Giga-Scale Enumerated Chemical Libraries.J Chem Inf Model. 2023 Sep 25;63(18):5773-5783. doi: 10.1021/acs.jcim.3c01239. Epub 2023 Sep 1. J Chem Inf Model. 2023. PMID: 37655823 Free PMC article.
References
-
- Willett P, Winterman V, Bawden D. Implementation of nearest-neighbor searching in an online chemical structure search system. J Chem Inf Comput Sci. 1986;26:36–41. doi: 10.1021/ci00049a008. - DOI
-
- Barnard JM, Downs GM. Clustering of chemical structures on the basis of two-dimensional similarity measures. J Chem Inf Comput Sci. 1992;32:644–649. doi: 10.1021/ci00010a010. - DOI
-
- Willett P, Barnard JM, Downs GM. Chemical similarity searching. J Chem Inf Comput Sci. 1998;38:983–996. doi: 10.1021/ci9800211. - DOI
-
- MACCS Structural Keys, Molecular Design Ltd., San Leandro, California, USA
LinkOut - more resources
Full Text Sources
Miscellaneous

