Molecular representations in AI-driven drug discovery: a review and practical guide
- PMID: 33431035
- PMCID: PMC7495975
- DOI: 10.1186/s13321-020-00460-5
Molecular representations in AI-driven drug discovery: a review and practical guide
Abstract
The technological advances of the past century, marked by the computer revolution and the advent of high-throughput screening technologies in drug discovery, opened the path to the computational analysis and visualization of bioactive molecules. For this purpose, it became necessary to represent molecules in a syntax that would be readable by computers and understandable by scientists of various fields. A large number of chemical representations have been developed over the years, their numerosity being due to the fast development of computers and the complexity of producing a representation that encompasses all structural and chemical characteristics. We present here some of the most popular electronic molecular and macromolecular representations used in drug discovery, many of which are based on graph representations. Furthermore, we describe applications of these representations in AI-driven drug discovery. Our aim is to provide a brief guide on structural representations that are essential to the practice of AI in drug discovery. This review serves as a guide for researchers who have little experience with the handling of chemical representations and plan to work on applications at the interface of these fields.
Keywords: Artificial intelligence; Cheminformatics; Drug discovery; Linear notation; Macromolecules; Molecular graphs; Molecular representation; Reaction prediction; Small molecules.
Conflict of interest statement
All the authors were employed by AstraZeneca and declare no competing interests.
Figures




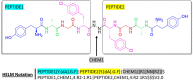

References
-
- Lawlor B. The chemical structure association trust. Chem Int. 2016;38(2):12–15. doi: 10.1515/ci-2016-0206. - DOI
-
- Wiswesser WJ. 107 years of line-formula notations (1861–968) J Chem Doc. 1968;8(3):146–150. doi: 10.1021/c160030a007. - DOI
-
- Zhou P, Shang Z. 2D molecular graphics: a flattened world of chemistry and biology - PubMed
-
- RasMol and OpenRasMol. http://www.openrasmol.org/. Accessed 27 Apr 2020.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

