PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF
- PMID: 33431576
- PMCID: PMC8666828
- DOI: 10.1136/gutjnl-2020-323395
PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF
Abstract
Objective: Hepatitis B virus-related acute-on-chronic liver failure (HBV-ACLF) pathophysiology remains unclear. This study aims to characterise the molecular basis of HBV-ACLF using transcriptomics.
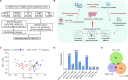
Methods: Four hundred subjects with HBV-ACLF, acute-on-chronic hepatic dysfunction (ACHD), liver cirrhosis (LC) or chronic hepatitis B (CHB) and normal controls (NC) from a prospective multicentre cohort were studied, and 65 subjects (ACLF, 20; ACHD, 10; LC, 10; CHB, 10; NC, 15) among them underwent mRNA sequencing using peripheral blood mononuclear cells (PBMCs).
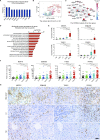
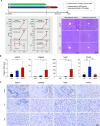
Results: The functional synergy analysis focusing on seven bioprocesses related to the PBMC response and the top 500 differentially expressed genes (DEGs) showed that viral processes were associated with all disease stages. Immune dysregulation, as the most prominent change and disorder triggered by HBV exacerbation, drove CHB or LC to ACHD and ACLF. Metabolic disruption was significant in ACHD and severe in ACLF. The analysis of 62 overlapping DEGs further linked the HBV-based immune-metabolism disorder to ACLF progression. The signatures of interferon-related, neutrophil-related and monocyte-related pathways related to the innate immune response were significantly upregulated. Signatures linked to the adaptive immune response were downregulated. Disruptions of lipid and fatty acid metabolism were observed during ACLF development. External validation of four DEGs underlying the aforementioned molecular mechanism in patients and experimental rats confirmed their specificity and potential as biomarkers for HBV-ACLF pathogenesis.
Conclusions: This study highlights immune-metabolism disorder triggered by HBV exacerbation as a potential mechanism of HBV-ACLF and may indicate a novel diagnostic and treatment target to reduce HBV-ACLF-related mortality.
Keywords: cirrhosis; hepatitis B; liver failure.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
