CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis
- PMID: 33431788
- PMCID: PMC7801454
- DOI: 10.1038/s41392-020-00437-8
CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis
Abstract
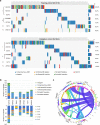
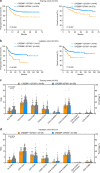
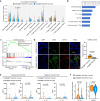
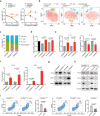
Epigenetic alterations play an important role in tumor progression of diffuse large B-cell lymphoma (DLBCL). However, the biological relevance of epigenetic gene mutations on tumor microenvironment remains to be determined. The core set of genes relating to histone methylation (KMT2D, KMT2C, EZH2), histone acetylation (CREBBP, EP300), DNA methylation (TET2), and chromatin remodeling (ARID1A) were detected in the training cohort of 316 patients by whole-genome/exome sequencing (WGS/WES) and in the validation cohort of 303 patients with newly diagnosed DLBCL by targeted sequencing. Their correlation with peripheral blood immune cells and clinical outcomes were assessed. Underlying mechanisms on tumor microenvironment were investigated both in vitro and in vivo. Among all 619 DLBCL patients, somatic mutations in KMT2D (19.5%) were most frequently observed, followed by mutations in ARID1A (8.7%), CREBBP (8.4%), KMT2C (8.2%), TET2 (7.8%), EP300 (6.8%), and EZH2 (2.9%). Among them, CREBBP/EP300 mutations were significantly associated with decreased peripheral blood absolute lymphocyte-to-monocyte ratios, as well as inferior progression-free and overall survival. In B-lymphoma cells, the mutation or knockdown of CREBBP or EP300 inhibited H3K27 acetylation, downregulated FBXW7 expression, activated the NOTCH pathway, and downstream CCL2/CSF1 expression, resulting in tumor-associated macrophage polarization to M2 phenotype and tumor cell proliferation. In B-lymphoma murine models, xenografted tumors bearing CREBBP/EP300 mutation presented lower H3K27 acetylation, higher M2 macrophage recruitment, and more rapid tumor growth than those with CREBBP/EP300 wild-type control via FBXW7-NOTCH-CCL2/CSF1 axis. Our work thus contributed to the understanding of aberrant histone acetylation regulation on tumor microenvironment as an alternative mechanism of tumor progression in DLBCL.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Coiffier B, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116:2040–2045. doi: 10.1182/blood-2010-03-276246. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

