HERC2 inactivation abrogates nucleolar localization of RecQ helicases BLM and WRN
- PMID: 33432007
- PMCID: PMC7801386
- DOI: 10.1038/s41598-020-79715-y
HERC2 inactivation abrogates nucleolar localization of RecQ helicases BLM and WRN
Abstract
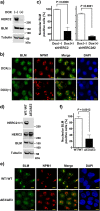
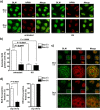
The nucleolus is a nuclear structure composed of ribosomal DNA (rDNA), and functions as a site for rRNA synthesis and processing. The rDNA is guanine-rich and prone to form G-quadruplex (G4), a secondary structure of DNA. We have recently found that HERC2, an HECT ubiquitin ligase, promotes BLM and WRN RecQ DNA helicases to resolve the G4 structure. Here, we report the role of HERC2 in the regulation of nucleolar localization of the helicases. Furthermore, HERC2 inactivation enhances the effects of CX-5461, an inhibitor of RNA polymerase I (Pol I)-mediated transcription of rRNA with an intrinsic G4-stabilizing activity. HERC2 depletion or homozygous deletion of the C-terminal HECT domain of HERC2 prevented the nucleolar localization of BLM and WRN, and inhibited relocalization of BLM to replication stress-induced nuclear RPA foci. HERC2 colocalized with fibrillarin and Pol I subunit RPA194, both of which are required for rRNA transcription. The HERC2 dysfunction enhanced the suppression of pre-rRNA transcription by CX-5461. These results suggest the effect of HERC2 status on the functions of BLM and WRN on rRNA transcription in the nucleolus. Since HERC2 is downregulated in numerous cancers, this effect may be clinically relevant considering the beneficial effects of CX-5461 in cancer treatments.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

