Modeling human adaptive immune responses with tonsil organoids
- PMID: 33432170
- PMCID: PMC7891554
- DOI: 10.1038/s41591-020-01145-0
Modeling human adaptive immune responses with tonsil organoids
Abstract
Most of what we know about adaptive immunity has come from inbred mouse studies, using methods that are often difficult or impossible to confirm in humans. In addition, vaccine responses in mice are often poorly predictive of responses to those same vaccines in humans. Here we use human tonsils, readily available lymphoid organs, to develop a functional organotypic system that recapitulates key germinal center features in vitro, including the production of antigen-specific antibodies, somatic hypermutation and affinity maturation, plasmablast differentiation and class-switch recombination. We use this system to define the essential cellular components necessary to produce an influenza vaccine response. We also show that it can be used to evaluate humoral immune responses to two priming antigens, rabies vaccine and an adenovirus-based severe acute respiratory syndrome coronavirus 2 vaccine, and to assess the effects of different adjuvants. This system should prove useful for studying critical mechanisms underlying adaptive immunity in much greater depth than previously possible and to rapidly test vaccine candidates and adjuvants in an entirely human system.
Conflict of interest statement
Competing interests
S.N.T., M.C. and E.G.D. are employed and have stock options with the biotechnology company Vaxart. P.L.F. and D.H.D. have shares in Nanommune, a company that uses Sino Biological’s proteins on commercially available protein microarrays. L.E.W., A.S., C.M.C., B.S.W., M.M.L., V.M., L.P.J., J.Z.A., L.K.B., N.G., K.J.L.J., F.Y., K.R., K.M.R., K.M.B., K.D.M., I.N.A., A.I.S., A.J., G.B.H., P.S.K., W.H.R., S.D.B., C.J.K. and M.M.D. declare no competing interests.
Figures

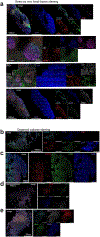
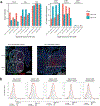

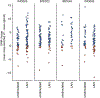
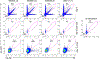








Comment in
-
Human tonsils in a dish.Nat Methods. 2021 Mar;18(3):229. doi: 10.1038/s41592-021-01093-8. Nat Methods. 2021. PMID: 33674799 No abstract available.
-
Tonsil organoids: peering down the throat of human immunity.Trends Immunol. 2021 May;42(5):367-368. doi: 10.1016/j.it.2021.03.009. Epub 2021 Mar 30. Trends Immunol. 2021. PMID: 33795204
References
-
- Behring E Untersuchungen uber das Zustandekommen der Diphtherie-Immunitat bei Thieren. Dtsch. Med. Wschr 16, 1145–1147 (1890).
-
- Behring E & Kitasato S Ueber das Zustandekommen der Diphtherie-Immunitat und der Tetanus-Immunitat bei Thieren. Dtsch. Med. Wschr 16, 1113–1114 (1890).
-
- Kaufmann SHE Emil von Behring: translational medicine at the dawn of immunology. Nat. Rev. Immunol 17, 341–343 (2017). - PubMed
-
- Kaufmann SH Immunology’s foundation: the 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nat. Immunol 9, 705–712 (2008). - PubMed
-
- Miller JF The discovery of thymus function and of thymus-derived lymphocytes. Immunol. Rev 185, 7–14 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 CA224081/CA/NCI NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U24 DK085532/DK/NIDDK NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
- R01 AI127877/AI/NIAID NIH HHS/United States
- R01 AI130398/AI/NIAID NIH HHS/United States
- S10 RR025518/RR/NCRR NIH HHS/United States
- K08 DE027730/DE/NIDCR NIH HHS/United States
- U01 DK085527/DK/NIDDK NIH HHS/United States
- U01 CA217851/CA/NCI NIH HHS/United States
- U19 AI116484/AI/NIAID NIH HHS/United States
- S10 RR027431/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

