The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing
- PMID: 33432195
- PMCID: PMC7983596
- DOI: 10.1038/s41593-020-00765-6
The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing
Erratum in
-
Author Correction: The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing.Nat Neurosci. 2021 Apr;24(4):611. doi: 10.1038/s41593-021-00830-8. Nat Neurosci. 2021. PMID: 33753946 Free PMC article. No abstract available.
-
Author Correction: The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing.Nat Neurosci. 2023 Oct;26(10):1833. doi: 10.1038/s41593-023-01437-x. Nat Neurosci. 2023. PMID: 37644260 No abstract available.
Abstract
We characterize the landscape of somatic mutations-mutations occurring after fertilization-in the human brain using ultra-deep (~250×) whole-genome sequencing of prefrontal cortex from 59 donors with autism spectrum disorder (ASD) and 15 control donors. We observe a mean of 26 somatic single-nucleotide variants per brain present in ≥4% of cells, with enrichment of mutations in coding and putative regulatory regions. Our analysis reveals that the first cell division after fertilization produces ~3.4 mutations, followed by 2-3 mutations in subsequent generations. This suggests that a typical individual possesses ~80 somatic single-nucleotide variants present in ≥2% of cells-comparable to the number of de novo germline mutations per generation-with about half of individuals having at least one potentially function-altering somatic mutation somewhere in the cortex. ASD brains show an excess of somatic mutations in neural enhancer sequences compared with controls, suggesting that mosaic enhancer mutations may contribute to ASD risk.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
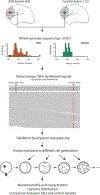
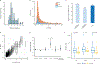

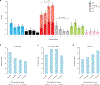

Comment in
-
Linking newly occurring mutations to autism.Nat Rev Genet. 2021 Mar;22(3):133. doi: 10.1038/s41576-021-00335-x. Nat Rev Genet. 2021. PMID: 33542502 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS032457/NS/NINDS NIH HHS/United States
- U01 MH106883/MH/NIMH NIH HHS/United States
- P50MH106933/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- U01 MH106876/MH/NIMH NIH HHS/United States
- T32GM007753/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- U54 HD090255/HD/NICHD NIH HHS/United States
- U01 MH106874/MH/NIMH NIH HHS/United States
- P50 MH106933/MH/NIMH NIH HHS/United States
- T32HG002295/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- R01NS032457/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- U01 MH106892/MH/NIMH NIH HHS/United States
- U01 MH108898/MH/NIMH NIH HHS/United States
- F31 MH124393/MH/NIMH NIH HHS/United States
- U01 MH106884/MH/NIMH NIH HHS/United States
- P50 HD103538/HD/NICHD NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- U01 MH106891/MH/NIMH NIH HHS/United States
- U01 MH106882/MH/NIMH NIH HHS/United States
- U01 MH106893/MH/NIMH NIH HHS/United States
- U01MH106883/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- T32 GM007544/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

