Exploring the Evolutionary History of Kinetic Stability in the α-Lytic Protease Family
- PMID: 33433210
- PMCID: PMC8174401
- DOI: 10.1021/acs.biochem.0c00720
Exploring the Evolutionary History of Kinetic Stability in the α-Lytic Protease Family
Abstract
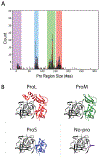
In addition to encoding the tertiary fold and stability, the primary sequence of a protein encodes the folding trajectory and kinetic barriers that determine the speed of folding. How these kinetic barriers are encoded is not well understood. Here, we use evolutionary sequence variation in the α-lytic protease (αLP) protein family to probe the relationship between sequence and energy landscape. αLP has an unusual energy landscape: the native state of αLP is not the most thermodynamically favored conformation and, instead, remains folded due to a large kinetic barrier preventing unfolding. To fold, αLP utilizes an N-terminal pro region similar in size to the protease itself that functions as a folding catalyst. Once folded, the pro region is removed, and the native state does not unfold on a biologically relevant time scale. Without the pro region, αLP folds on the order of millennia. A phylogenetic search uncovers αLP homologs with a wide range of pro region sizes, including some with no pro region at all. In the resulting phylogenetic tree, these homologs cluster by pro region size. By studying homologs naturally lacking a pro region, we demonstrate they can be thermodynamically stable, fold much faster than αLP, yet retain the same fold as αLP. Key amino acids thought to contribute to αLP's extreme kinetic stability are lost in these homologs, supporting their role in kinetic stability. This study highlights how the entire energy landscape plays an important role in determining the evolutionary pressures on the protein sequence.
Figures

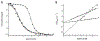


Similar articles
-
The folding landscape of an alpha-lytic protease variant reveals the role of a conserved beta-hairpin in the development of kinetic stability.Proteins. 2005 Oct 1;61(1):105-14. doi: 10.1002/prot.20525. Proteins. 2005. PMID: 16044461
-
Interdependent folding of the N- and C-terminal domains defines the cooperative folding of alpha-lytic protease.Biochemistry. 2003 Nov 18;42(45):13212-9. doi: 10.1021/bi035409q. Biochemistry. 2003. PMID: 14609332
-
Energetic landscape of alpha-lytic protease optimizes longevity through kinetic stability.Nature. 2002 Jan 17;415(6869):343-6. doi: 10.1038/415343a. Nature. 2002. PMID: 11797014
-
How do proteins avoid becoming too stable? Biophysical studies into metastable proteins.Eur Biophys J. 2004 Apr;33(2):83-8. doi: 10.1007/s00249-003-0356-1. Epub 2003 Sep 19. Eur Biophys J. 2004. PMID: 14504841 Review.
-
Common themes and variations in serine protease autotransporters.Trends Microbiol. 2008 Aug;16(8):370-9. doi: 10.1016/j.tim.2008.05.003. Epub 2008 Jul 1. Trends Microbiol. 2008. PMID: 18595714 Review.
Cited by
-
Ancestral Reconstruction and the Evolution of Protein Energy Landscapes.Annu Rev Biophys. 2024 Jul;53(1):127-146. doi: 10.1146/annurev-biophys-030722-125440. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38134334 Free PMC article. Review.
-
Exploring the sequence and structural determinants of the energy landscape from thermodynamically stable and kinetically trapped subtilisins: ISP1 and SbtE.bioRxiv [Preprint]. 2025 Feb 26:2024.09.08.611919. doi: 10.1101/2024.09.08.611919. bioRxiv. 2025. Update in: Protein Sci. 2025 Sep;34(9):e70264. doi: 10.1002/pro.70264. PMID: 39314365 Free PMC article. Updated. Preprint.
-
Quantifying protein unfolding kinetics with a high-throughput microfluidic platform.bioRxiv [Preprint]. 2025 Jan 18:2025.01.15.633299. doi: 10.1101/2025.01.15.633299. bioRxiv. 2025. PMID: 39868203 Free PMC article. Preprint.
-
Allelic variations in the chpG effector gene within Clavibacter michiganensis populations determine pathogen host range.PLoS Pathog. 2024 Jul 19;20(7):e1012380. doi: 10.1371/journal.ppat.1012380. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39028765 Free PMC article.
-
The conformational landscape of a serpin N-terminal subdomain facilitates folding and in-cell quality control.bioRxiv [Preprint]. 2023 Apr 26:2023.04.24.537978. doi: 10.1101/2023.04.24.537978. bioRxiv. 2023. PMID: 37163105 Free PMC article. Preprint.
References
-
- Jaenicke R; Böhm G (1998) The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8, 738–748. - PubMed
-
- Colón W; Church J; Sen J; Thibeault J; Trasatti H; Xia K (2017) Biological Roles of Protein Kinetic Stability. Biochemistry 56, 6179–6186. - PubMed
-
- Sohl JL; Jaswal SS; Agard DA (1998) Unfolded conformations of α-lytic protease are more stable than its native state. Nature 395, 817–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

