PARP-mediated PARylation of MGMT is critical to promote repair of temozolomide-induced O6-methylguanine DNA damage in glioblastoma
- PMID: 33433610
- PMCID: PMC8168825
- DOI: 10.1093/neuonc/noab003
PARP-mediated PARylation of MGMT is critical to promote repair of temozolomide-induced O6-methylguanine DNA damage in glioblastoma
Abstract
Background: Temozolomide (TMZ) resistance in glioblastoma multiforme (GBM) is mediated by the DNA repair protein O6-methylguanine DNA methyltransferase (MGMT). MGMT promoter methylation (occurs in about 40% of patients) is associated with loss of MGMT expression (MGMT-) that compromises DNA repair, leading to a favorable response to TMZ therapy. The 60% of patients with unmethylated MGMT (MGMT+) GBM experience resistance to TMZ; in these patients, understanding the mechanism of MGMT-mediated repair and modulating MGMT activity may lead to enhanced TMZ activity. Here, we report a novel mode of regulation of MGMT protein activity by poly(ADP-ribose) polymerase (PARP).
Methods: MGMT-PARP interaction was detected by co-immunoprecipitation. PARylation of MGMT and PARP was detected by co-immunoprecipitation with anti-PAR antibody. O6-methylguanine (O6-MetG) adducts were quantified by immunofluorescence assay. In vivo studies were conducted in mice to determine the effectiveness of PARP inhibition in sensitizing GBM to TMZ.
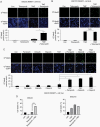
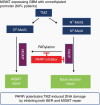
Results: We demonstrated that PARP physically binds with MGMT and PARylates MGMT in response to TMZ treatment. In addition, PARylation of MGMT by PARP is required for MGMT binding to chromatin to enhance the removal of O6-MetG adducts from DNA after TMZ treatment. PARP inhibitors reduced PARP-MGMT binding and MGMT PARylation, silencing MGMT activity to repair O6-MetG. PARP inhibition restored TMZ sensitivity in vivo in MGMT-expressing GBM.
Conclusion: This study demonstrated that PARylation of MGMT by PARP is critical for repairing TMZ-induced O6-MetG, and inhibition of PARylation by PARP inhibitor reduces MGMT function rendering sensitization to TMZ, providing a rationale for combining PARP inhibitors to sensitize TMZ in MGMT-unmethylated GBM.
Keywords: DNA damage repair; MGMT PARylation; PARP; TMZ resistance.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures




Similar articles
-
Temozolomide: mechanisms of action, repair and resistance.Curr Mol Pharmacol. 2012 Jan;5(1):102-14. doi: 10.2174/1874467211205010102. Curr Mol Pharmacol. 2012. PMID: 22122467
-
Delineation of MGMT Hypermethylation as a Biomarker for Veliparib-Mediated Temozolomide-Sensitizing Therapy of Glioblastoma.J Natl Cancer Inst. 2015 Nov 27;108(5):djv369. doi: 10.1093/jnci/djv369. Print 2016 May. J Natl Cancer Inst. 2015. PMID: 26615020 Free PMC article.
-
A novel compound EPIC-0412 reverses temozolomide resistance via inhibiting DNA repair/MGMT in glioblastoma.Neuro Oncol. 2023 May 4;25(5):857-870. doi: 10.1093/neuonc/noac242. Neuro Oncol. 2023. PMID: 36272139 Free PMC article.
-
Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review).Int J Oncol. 2015 Aug;47(2):417-28. doi: 10.3892/ijo.2015.3026. Epub 2015 May 29. Int J Oncol. 2015. PMID: 26035292 Free PMC article. Review.
-
Unlocking temozolomide resistance in glioblastoma: the pivotal role of MicroRNAs and in-silico insights.Med Oncol. 2025 Jul 17;42(8):343. doi: 10.1007/s12032-025-02884-1. Med Oncol. 2025. PMID: 40676394 Review.
Cited by
-
Advances in medical treatment for pancreatic neuroendocrine neoplasms.World J Gastroenterol. 2022 May 28;28(20):2163-2175. doi: 10.3748/wjg.v28.i20.2163. World J Gastroenterol. 2022. PMID: 35721885 Free PMC article. Review.
-
[Curcumol reverses temozolomide resistance in glioma cells by regulating the UTX/MGMT axis].Nan Fang Yi Ke Da Xue Xue Bao. 2023 Oct 20;43(10):1697-1705. doi: 10.12122/j.issn.1673-4254.2023.10.07. Nan Fang Yi Ke Da Xue Xue Bao. 2023. PMID: 37933645 Free PMC article. Chinese.
-
Revisiting Temozolomide's role in solid tumors: Old is gold?J Cancer. 2024 Apr 22;15(11):3254-3271. doi: 10.7150/jca.94109. eCollection 2024. J Cancer. 2024. PMID: 38817857 Free PMC article. Review.
-
Radiomics for predicting MGMT status in cerebral glioblastoma: comparison of different MRI sequences.J Radiat Res. 2024 May 23;65(3):350-359. doi: 10.1093/jrr/rrae007. J Radiat Res. 2024. PMID: 38650477 Free PMC article.
-
HOXD-AS2-STAT3 feedback loop attenuates sensitivity to temozolomide in glioblastoma.CNS Neurosci Ther. 2023 Nov;29(11):3430-3445. doi: 10.1111/cns.14277. Epub 2023 Jun 12. CNS Neurosci Ther. 2023. PMID: 37308741 Free PMC article.
References
-
- Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. - PubMed
-
- Esteller M, Garcia-Foncillas J, Andion E, et al. . Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. - PubMed
-
- Palma JP, Rodriguez LE, Bontcheva-Diaz VD, et al. . The PARP inhibitor, ABT-888 potentiates temozolomide: correlation with drug levels and reduction in PARP activity in vivo. Anticancer Res. 2008;28(5A): 2625–2635. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

