Neuroinvasion of SARS-CoV-2 in human and mouse brain
- PMID: 33433624
- PMCID: PMC7808299
- DOI: 10.1084/jem.20202135
Neuroinvasion of SARS-CoV-2 in human and mouse brain
Abstract
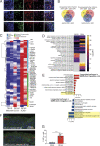
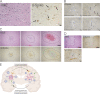
Although COVID-19 is considered to be primarily a respiratory disease, SARS-CoV-2 affects multiple organ systems including the central nervous system (CNS). Yet, there is no consensus on the consequences of CNS infections. Here, we used three independent approaches to probe the capacity of SARS-CoV-2 to infect the brain. First, using human brain organoids, we observed clear evidence of infection with accompanying metabolic changes in infected and neighboring neurons. However, no evidence for type I interferon responses was detected. We demonstrate that neuronal infection can be prevented by blocking ACE2 with antibodies or by administering cerebrospinal fluid from a COVID-19 patient. Second, using mice overexpressing human ACE2, we demonstrate SARS-CoV-2 neuroinvasion in vivo. Finally, in autopsies from patients who died of COVID-19, we detect SARS-CoV-2 in cortical neurons and note pathological features associated with infection with minimal immune cell infiltrates. These results provide evidence for the neuroinvasive capacity of SARS-CoV-2 and an unexpected consequence of direct infection of neurons by SARS-CoV-2.
© 2021 Song et al.
Conflict of interest statement
Disclosures: M. Gunel reported personal fees from AI Therapeutics outside the submitted work; and reported, "AI Therapeutics is currently sponsoring a clinical trial for a therapeutic, which has no relevance for this study, in COVID-19. I am the Chief Scientific Advisor to AI Therapeutics." C.B. Wilen reported personal fees from ZymoResearch outside the submitted work; in addition, C.B. Wilen had a patent for compounds and compositions for treating, ameliorating, and/or preventing SARS-CoV-2 infection and/or complications thereof pending. S. Haik reported a patent to Method for treating prion diseases (PCT/EP 2019/070457) pending. A. Iwasaki reported "other" from RIGImmune and grants from Spring Discovery during the conduct of the study; in addition, A. Iwasaki had a patent to 14/776,463 pending, a patent for a T cell-based immunotherapy for central nervous system viral infections and tumors pending, and a patent to manipulation of meningeal lymphatic vasculature for brain and CNS tumor therapy pending. No other disclosures were reported.
Figures








Update of
-
Neuroinvasion of SARS-CoV-2 in human and mouse brain.bioRxiv [Preprint]. 2020 Sep 8:2020.06.25.169946. doi: 10.1101/2020.06.25.169946. bioRxiv. 2020. Update in: J Exp Med. 2021 Mar 1;218(3):e20202135. doi: 10.1084/jem.20202135. PMID: 32935108 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

