Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]
- PMID: 33434234
- PMCID: PMC7802970
- DOI: 10.1371/journal.pone.0244593
Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]
Abstract
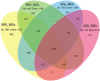
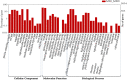
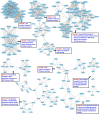
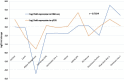
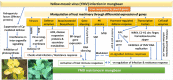
Yellow Mosaic Disease (YMD) in mungbean [Vigna radiata (L.) R. Wilczek] is one of the most damaging diseases in Asia. In the northern part of India, the YMD is caused by Mungbean Yellow Mosaic India Virus (MYMIV), while in southern India this is caused by Mungbean Yellow Mosaic Virus (MYMV). The molecular mechanism of YMD resistance in mungbean remains largely unknown. In this study, RNA-seq analysis was conducted between a resistant (PMR-1) and a susceptible (Pusa Vishal) mungbean genotype under infected and control conditions to understand the regulatory network operating between mungbean-YMV. Overall, 76.8 million raw reads could be generated in different treatment combinations, while mapping rate per library to the reference genome varied from 86.78% to 93.35%. The resistance to MYMIV showed a very complicated gene network, which begins with the production of general PAMPs (pathogen-associated molecular patterns), then activation of various signaling cascades like kinases, jasmonic acid (JA) and brassinosteroid (BR), and finally the expression of specific genes (like PR-proteins, virus resistance and R-gene proteins) leading to resistance response. The function of WRKY, NAC and MYB transcription factors in imparting the resistance against MYMIV could be established. The string analysis also revealed the role of proteins involved in kinase, viral movement and phytoene synthase activity in imparting YMD resistance. A set of novel stress-related EST-SSRs are also identified from the RNA-Seq data which may be used to find the linked genes/QTLs with the YMD resistance. Also, 11 defence-related transcripts could be validated through quantitative real-time PCR analysis. The identified gene networks have led to an insight about the defence mechanism operating against MYMIV infection in mungbean which will be of immense use to manage the YMD resistance in mungbean.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Jain HK, Mehra KL (1980) Evaluation, adaptation, relationship and cases of the species of Vigna cultivation in Asia. In: Summerfield RJ, Butnting AH(eds) Advances in legume science. Royal Botanical Garden, Kew, London, vol 273, pp 459–468. 10.1017/S0014479700011327 - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

