Multiple Alu Exonization in 3'UTR of a Primate-Specific Isoform of CYP20A1 Creates a Potential miRNA Sponge
- PMID: 33434274
- PMCID: PMC7802813
- DOI: 10.1093/gbe/evaa233
Multiple Alu Exonization in 3'UTR of a Primate-Specific Isoform of CYP20A1 Creates a Potential miRNA Sponge
Abstract
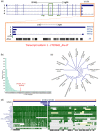
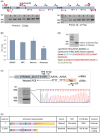
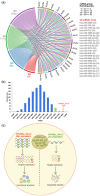
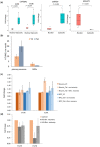
Alu repeats contribute to phylogenetic novelties in conserved regulatory networks in primates. Our study highlights how exonized Alus could nucleate large-scale mRNA-miRNA interactions. Using a functional genomics approach, we characterize a transcript isoform of an orphan gene, CYP20A1 (CYP20A1_Alu-LT) that has exonization of 23 Alus in its 3'UTR. CYP20A1_Alu-LT, confirmed by 3'RACE, is an outlier in length (9 kb 3'UTR) and widely expressed. Using publically available data sets, we demonstrate its expression in higher primates and presence in single nucleus RNA-seq of 15,928 human cortical neurons. miRanda predicts ∼4,700 miRNA recognition elements (MREs) for ∼1,000 miRNAs, primarily originated within these 3'UTR-Alus. CYP20A1_Alu-LT could be a potential multi-miRNA sponge as it harbors ≥10 MREs for 140 miRNAs and has cytosolic localization. We further tested whether expression of CYP20A1_Alu-LT correlates with mRNAs harboring similar MRE targets. RNA-seq with conjoint miRNA-seq analysis was done in primary human neurons where we observed CYP20A1_Alu-LT to be downregulated during heat shock response and upregulated in HIV1-Tat treatment. In total, 380 genes were positively correlated with its expression (significantly downregulated in heat shock and upregulated in Tat) and they harbored MREs for nine expressed miRNAs which were also enriched in CYP20A1_Alu-LT. MREs were significantly enriched in these 380 genes compared with random sets of differentially expressed genes (P = 8.134e-12). Gene ontology suggested involvement of these genes in neuronal development and hemostasis pathways thus proposing a novel component of Alu-miRNA-mediated transcriptional modulation that could govern specific physiological outcomes in higher primates.
Keywords: 3 prime UnTranslated Region (3′UTR) extension; Alu-miRNA; Cytochrome P450 20A1 (CYP20A1); miRNA recognition elements (MREs); multi-miRNA sponge; neurocoagulopathy.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Exonized Alu repeats in the 3'UTR of a CYP20A1_Alu-LT transcript act as a miRNA sponge.BMC Res Notes. 2023 Mar 9;16(1):32. doi: 10.1186/s13104-023-06289-z. BMC Res Notes. 2023. PMID: 36895043 Free PMC article.
-
Alu-miRNA interactions modulate transcript isoform diversity in stress response and reveal signatures of positive selection.Sci Rep. 2016 Sep 2;6:32348. doi: 10.1038/srep32348. Sci Rep. 2016. PMID: 27586304 Free PMC article.
-
Novel Role of 3'UTR-Embedded Alu Elements as Facilitators of Processed Pseudogene Genesis and Host Gene Capture by Viral Genomes.PLoS One. 2016 Dec 29;11(12):e0169196. doi: 10.1371/journal.pone.0169196. eCollection 2016. PLoS One. 2016. PMID: 28033411 Free PMC article.
-
From 'JUNK' to just unexplored noncoding knowledge: the case of transcribed Alus.Brief Funct Genomics. 2011 Sep;10(5):294-311. doi: 10.1093/bfgp/elr029. Brief Funct Genomics. 2011. PMID: 21987713 Review.
-
The role of Alu elements in the cis-regulation of RNA processing.Cell Mol Life Sci. 2015 Nov;72(21):4063-76. doi: 10.1007/s00018-015-1990-3. Epub 2015 Jul 30. Cell Mol Life Sci. 2015. PMID: 26223268 Free PMC article. Review.
Cited by
-
More than causing (epi)genomic instability: emerging physiological implications of transposable element modulation.J Biomed Sci. 2021 Aug 7;28(1):58. doi: 10.1186/s12929-021-00754-2. J Biomed Sci. 2021. PMID: 34364371 Free PMC article. Review.
-
Retrotransposons as Drivers of Mammalian Brain Evolution.Life (Basel). 2021 Apr 22;11(5):376. doi: 10.3390/life11050376. Life (Basel). 2021. PMID: 33922141 Free PMC article. Review.
-
PRRGO: A Tool for Visualizing and Mapping Globally Expressed Genes in Public Gene Expression Omnibus RNA-Sequencing Studies to PageRank-scored Gene Ontology Terms.bioRxiv [Preprint]. 2024 Jan 24:2024.01.21.576540. doi: 10.1101/2024.01.21.576540. bioRxiv. 2024. PMID: 38328158 Free PMC article. Preprint.
-
The Role of Transposable Elements of the Human Genome in Neuronal Function and Pathology.Int J Mol Sci. 2022 May 23;23(10):5847. doi: 10.3390/ijms23105847. Int J Mol Sci. 2022. PMID: 35628657 Free PMC article. Review.
-
miR-17-5p-Mediated RNA Activation Upregulates KPNA2 Expression and Inhibits High-Glucose-Induced Apoptosis of Sheep Granulosa Cells.Int J Mol Sci. 2025 Jan 23;26(3):943. doi: 10.3390/ijms26030943. Int J Mol Sci. 2025. PMID: 39940713 Free PMC article.
References
-
- Batzer MA, Deininger PL. 1991. A human-specific subfamily of Alu sequences. Genomics 9(3):481–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

