A Set of Cell Lines Derived from a Genetic Murine Glioblastoma Model Recapitulates Molecular and Morphological Characteristics of Human Tumors
- PMID: 33435218
- PMCID: PMC7827614
- DOI: 10.3390/cancers13020230
A Set of Cell Lines Derived from a Genetic Murine Glioblastoma Model Recapitulates Molecular and Morphological Characteristics of Human Tumors
Abstract
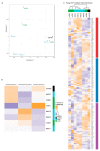
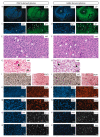
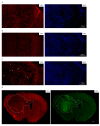
Glioblastomas (GBM) are the most aggressive tumors affecting the central nervous system in adults, causing death within, on average, 15 months after diagnosis. Immunocompetent in-vivo models that closely mirror human GBM are urgently needed for deciphering glioma biology and for the development of effective treatment options. The murine GBM cell lines currently available for engraftment in immunocompetent mice are not only exiguous but also inadequate in representing prominent characteristics of human GBM such as infiltrative behavior, necrotic areas, and pronounced tumor heterogeneity. Therefore, we generated a set of glioblastoma cell lines by repeated in vivo passaging of cells isolated from a neural stem cell-specific Pten/p53 double-knockout genetic mouse brain tumor model. Transcriptome and genome analyses of the cell lines revealed molecular heterogeneity comparable to that observed in human glioblastoma. Upon orthotopic transplantation into syngeneic hosts, they formed high-grade gliomas that faithfully recapitulated the histopathological features, invasiveness and immune cell infiltration characteristic of human glioblastoma. These features make our cell lines unique and useful tools to study multiple aspects of glioblastoma pathomechanism and to test novel treatments in an intact immune microenvironment.
Keywords: glioblastoma; mouse model; syngeneic cell line.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Weller M., van den Bent M., Tonn J.C., Stupp R., Preusser M., Cohen-Jonathan-Moyal E., Henriksson R., Le Rhun E., Balana C., Chinot O., et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315–e329. doi: 10.1016/S1470-2045(17)30194-8. - DOI - PubMed
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
-
- Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., deCarvalho A.C., Lyu S., Li P., Li Y., et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

