Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA
- PMID: 33435235
- PMCID: PMC7827361
- DOI: 10.3390/insects12010049
Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA
Abstract
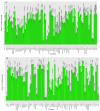
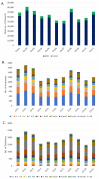
The production and quality of Phaseolus vulgaris (snap bean) have been negatively impacted by leaf crumple disease caused by two whitefly-transmitted begomoviruses: cucurbit leaf crumple virus (CuLCrV) and sida golden mosaic Florida virus (SiGMFV), which often appear as a mixed infection in Georgia. Host resistance is the most economical management strategy against whitefly-transmitted viruses. Currently, information is not available with respect to resistance to these two viruses in commercial cultivars. In two field seasons (2018 and 2019), we screened Phaseolus spp. genotypes (n = 84 in 2018; n = 80 in 2019; most of the genotypes were common in both years with a few exceptions) for resistance against CuLCrV and/or SiGMFV. We also included two commonly grown Lima bean (Phaseolus lunatus) varieties in our field screening. Twenty Phaseolus spp. genotypes with high to moderate-levels of resistance (disease severity ranging from 5%-50%) to CuLCrV and/or SiGMFV were identified. Twenty-one Phaseolus spp. genotypes were found to be highly susceptible with a disease severity of ≥66%. Furthermore, based on the greenhouse evaluation with two genotypes-each (two susceptible and two resistant; identified in field screen) exposed to viruliferous whiteflies infected with CuLCrV and SiGMFV, we observed that the susceptible genotypes accumulated higher copy numbers of both viruses and displayed severe crumple severity compared to the resistant genotypes, indicating that resistance might potentially be against the virus complex rather than against the whiteflies. Adult whitefly counts differed significantly among Phaseolus genotypes in both years. The whole genome of these Phaseolus spp. [snap bean (n = 82); Lima bean (n = 2)] genotypes was sequenced and genetic variability among them was identified. Over 900 giga-base (Gb) of filtered data were generated and >88% of the resulting data were mapped to the reference genome, and SNP and Indel variants in Phaseolus spp. genotypes were obtained. A total of 645,729 SNPs and 68,713 Indels, including 30,169 insertions and 38,543 deletions, were identified, which were distributed in 11 chromosomes with chromosome 02 harboring the maximum number of variants. This phenotypic and genotypic information will be helpful in genome-wide association studies that will aid in identifying the genetic basis of resistance to these begomoviruses in Phaseolus spp.
Keywords: cucurbit leaf crumple virus; lima beans; sida golden mosaic Florida virus; snap beans; whitefly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- United States Dry Bean Council. [(accessed on 20 November 2020)]; Available online: https://www.usdrybeans.com/industry/production-facts/
-
- Georgia Agricultural Commodity Rankings. [(accessed on 22 November 2020)];2018 Available online: https://caed.uga.edu/content/dam/caes-subsite/caed/publications/annual-r....
-
- Gautam S. The Role of Bemisia tabaci in the Transmission of Vegetable Viruses in the Farmscape of Georgia. University of Georgia; Athens, GA, USA: 2019.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

