The Roles of Chromatin Accessibility in Regulating the Candida albicans White-Opaque Phenotypic Switch
- PMID: 33435404
- PMCID: PMC7826875
- DOI: 10.3390/jof7010037
The Roles of Chromatin Accessibility in Regulating the Candida albicans White-Opaque Phenotypic Switch
Abstract
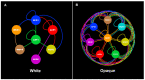
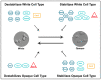
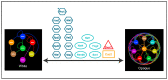
Candida albicans, a diploid polymorphic fungus, has evolved a unique heritable epigenetic program that enables reversible phenotypic switching between two cell types, referred to as "white" and "opaque". These cell types are established and maintained by distinct transcriptional programs that lead to differences in metabolic preferences, mating competencies, cellular morphologies, responses to environmental signals, interactions with the host innate immune system, and expression of approximately 20% of genes in the genome. Transcription factors (defined as sequence specific DNA-binding proteins) that regulate the establishment and heritable maintenance of the white and opaque cell types have been a primary focus of investigation in the field; however, other factors that impact chromatin accessibility, such as histone modifying enzymes, chromatin remodelers, and histone chaperone complexes, also modulate the dynamics of the white-opaque switch and have been much less studied to date. Overall, the white-opaque switch represents an attractive and relatively "simple" model system for understanding the logic and regulatory mechanisms by which heritable cell fate decisions are determined in higher eukaryotes. Here we review recent discoveries on the roles of chromatin accessibility in regulating the C. albicans white-opaque phenotypic switch.
Keywords: Candida albicans; cell fate decisions; chromatin; chromatin remodeling enzymes; epigenetics; heritability; histone chaperone complexes; histone modifying enzymes; transcriptional regulation; white-opaque switching.
Conflict of interest statement
The authors declare no conflict of interest pertaining to the topic of this manuscript.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

