Genetic Manipulation of the Brassicaceae Smut Fungus Thecaphora thlaspeos
- PMID: 33435409
- PMCID: PMC7826943
- DOI: 10.3390/jof7010038
Genetic Manipulation of the Brassicaceae Smut Fungus Thecaphora thlaspeos
Abstract
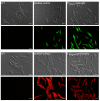
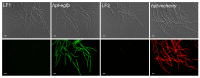
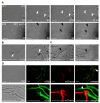
Investigation of plant-microbe interactions greatly benefit from genetically tractable partners to address, molecularly, the virulence and defense mechanisms. The smut fungus Ustilago maydis is a model pathogen in that sense: efficient homologous recombination and a small genome allow targeted modification. On the host side, maize is limiting with regard to rapid genetic alterations. By contrast, the model plant Arabidopsis thaliana is an excellent model with a vast amount of information and techniques as well as genetic resources. Here, we present a transformation protocol for the Brassicaceae smut fungus Thecaphora thlaspeos. Using the well-established methodology of protoplast transformation, we generated the first reporter strains expressing fluorescent proteins to follow mating. As a proof-of-principle for homologous recombination, we deleted the pheromone receptor pra1. As expected, this mutant cannot mate. Further analysis will contribute to our understanding of the role of mating for infection biology in this novel model fungus. From now on, the genetic manipulation of T. thlaspeos, which is able to colonize the model plant A. thaliana, provides us with a pathosystem in which both partners are genetically amenable to study smut infection biology.
Keywords: homologous recombination; infection; mating; pheromone receptor; protoplast; smut; transformation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Kronstad J.W. Pathogenesis and sexual development of the smut fungi. In: Stacey G.K.N.T., editor. Plant-Microbe Interactions. Volume 1 Springer; Boston, MA, USA: 1997.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

