Cell-Free DNA to Detect Heart Allograft Acute Rejection
- PMID: 33435695
- PMCID: PMC8221834
- DOI: 10.1161/CIRCULATIONAHA.120.049098
Cell-Free DNA to Detect Heart Allograft Acute Rejection
Abstract
Background: After heart transplantation, endomyocardial biopsy (EMBx) is used to monitor for acute rejection (AR). Unfortunately, EMBx is invasive, and its conventional histological interpretation has limitations. This is a validation study to assess the performance of a sensitive blood biomarker-percent donor-derived cell-free DNA (%ddcfDNA)-for detection of AR in cardiac transplant recipients.
Methods: This multicenter, prospective cohort study recruited heart transplant subjects and collected plasma samples contemporaneously with EMBx for %ddcfDNA measurement by shotgun sequencing. Histopathology data were collected to define AR, its 2 phenotypes (acute cellular rejection [ACR] and antibody-mediated rejection [AMR]), and controls without rejection. The primary analysis was to compare %ddcfDNA levels (median and interquartile range [IQR]) for AR, AMR, and ACR with controls and to determine %ddcfDNA test characteristics using receiver-operator characteristics analysis.
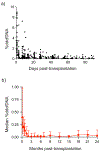
Results: The study included 171 subjects with median posttransplant follow-up of 17.7 months (IQR, 12.1-23.6), with 1392 EMBx, and 1834 %ddcfDNA measures available for analysis. Median %ddcfDNA levels decayed after surgery to 0.13% (IQR, 0.03%-0.21%) by 28 days. Also, %ddcfDNA increased again with AR compared with control values (0.38% [IQR, 0.31-0.83%], versus 0.03% [IQR, 0.01-0.14%]; P<0.001). The rise was detected 0.5 and 3.2 months before histopathologic diagnosis of ACR and AMR. The area under the receiver operator characteristic curve for AR was 0.92. A 0.25%ddcfDNA threshold had a negative predictive value for AR of 99% and would have safely eliminated 81% of EMBx. In addition, %ddcfDNA showed distinctive characteristics comparing AMR with ACR, including 5-fold higher levels (AMR ≥2, 1.68% [IQR, 0.49-2.79%] versus ACR grade ≥2R, 0.34% [IQR, 0.28-0.72%]), higher area under the receiver operator characteristic curve (0.95 versus 0.85), higher guanosine-cytosine content, and higher percentage of short ddcfDNA fragments.
Conclusions: We found that %ddcfDNA detected AR with a high area under the receiver operator characteristic curve and negative predictive value. Monitoring with ddcfDNA demonstrated excellent performance characteristics for both ACR and AMR and led to earlier detection than the EMBx-based monitoring. This study supports the use of %ddcfDNA to monitor for AR in patients with heart transplant and paves the way for a clinical utility study. Registration: URL: https://www.clinicaltrials.gov; Unique identifier: NCT02423070.
Keywords: allograft rejection; biomarkers; cardiac transplantation; cell-free DNA; graft injury.
Figures



Comment in
-
Solid Gold, or Liquid Gold?: Towards a New Diagnostic Standard for Heart Transplant Rejection.Circulation. 2021 Mar 23;143(12):1198-1201. doi: 10.1161/CIRCULATIONAHA.120.052925. Epub 2021 Mar 22. Circulation. 2021. PMID: 33750203 No abstract available.
-
Response by Shah et al to Letter Regarding Article, "Cell-Free DNA to Detect Heart Allograft Acute Rejection".Circulation. 2021 Sep 7;144(10):e198-e199. doi: 10.1161/CIRCULATIONAHA.121.055697. Epub 2021 Sep 7. Circulation. 2021. PMID: 34491771 No abstract available.
-
Letter by Amadio et al Regarding Article, "Cell-Free DNA to Detect Heart Allograft Acute Rejection".Circulation. 2021 Sep 7;144(10):e196-e197. doi: 10.1161/CIRCULATIONAHA.121.054333. Epub 2021 Sep 7. Circulation. 2021. PMID: 34491772 No abstract available.
Similar articles
-
Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study.J Heart Lung Transplant. 2021 Aug;40(8):822-830. doi: 10.1016/j.healun.2021.04.009. Epub 2021 Apr 24. J Heart Lung Transplant. 2021. PMID: 34130911 Free PMC article.
-
Donor-derived cell-free DNA for detection of acute rejection in lung transplant recipients.Front Immunol. 2025 Jan 29;16:1531774. doi: 10.3389/fimmu.2025.1531774. eCollection 2025. Front Immunol. 2025. PMID: 39944692 Free PMC article.
-
Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis.J Heart Lung Transplant. 2018 Jul;37(7):925-932. doi: 10.1016/j.healun.2018.01.1305. Epub 2018 Jan 31. J Heart Lung Transplant. 2018. PMID: 29500138
-
The Monitoring of Donor-derived Cell-free DNA in Kidney Transplantation.Transplantation. 2021 Mar 1;105(3):509-516. doi: 10.1097/TP.0000000000003393. Transplantation. 2021. PMID: 32732615 Review.
-
Analysis of the primary factors influencing donor derived cell-free DNA testing in kidney transplantation.Front Immunol. 2024 Sep 6;15:1435578. doi: 10.3389/fimmu.2024.1435578. eCollection 2024. Front Immunol. 2024. PMID: 39308855 Free PMC article. Review.
Cited by
-
Monitoring of plasma circulating donor DNA reflects cardiac graft injury: Report of two cases.Biomed Rep. 2024 Jan 29;20(3):50. doi: 10.3892/br.2024.1738. eCollection 2024 Mar. Biomed Rep. 2024. PMID: 38357233 Free PMC article.
-
Revisiting Biomarkers of Cardiac Allograft Vasculopathy: Addressing the Achilles Heel of Heart Transplantation.Curr Heart Fail Rep. 2024 Dec;21(6):580-590. doi: 10.1007/s11897-024-00685-7. Epub 2024 Oct 17. Curr Heart Fail Rep. 2024. PMID: 39414739 Review.
-
Electron Microscopy Reveals Evidence of Perinuclear Clustering of Mitochondria in Cardiac Biopsy-Proven Allograft Rejection.J Pers Med. 2022 Feb 17;12(2):296. doi: 10.3390/jpm12020296. J Pers Med. 2022. PMID: 35207783 Free PMC article.
-
Integrated cell-free DNA and cytokine analysis uncovers distinct tissue injury and immune response patterns in solid organ transplant recipients with COVID-19.Res Sq [Preprint]. 2022 Jan 20:rs.3.rs-1262270. doi: 10.21203/rs.3.rs-1262270/v1. Res Sq. 2022. PMID: 35075453 Free PMC article. Preprint.
-
Ten-Year Experience with Endomyocardial Biopsy after Orthotopic Heart Transplantation: Comparison between Trans-Jugular and Trans-Femoral Approach.J Cardiovasc Dev Dis. 2024 Apr 3;11(4):115. doi: 10.3390/jcdd11040115. J Cardiovasc Dev Dis. 2024. PMID: 38667732 Free PMC article.
References
-
- Mehra MR, Crespo-Leiro MG, Dipchand A, Ensminger SM, Hiemann NE, Kobashigawa JA, Madsen J, Parameshwar J, Starling RC and Uber PA. International Society for Heart and Lung Transplantation working formulation of a standardized nomenclature for cardiac allograft vasculopathy-2010. J Heart Lung Transplant. 2010;29:717–27. - PubMed
-
- Costanzo-Nordin MR. Cardiac allograft vasculopathy: relationship with acute cellular rejection and histocompatibility. J Heart Lung Transplant. 1992;11:S90–103. - PubMed
-
- Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, Andersen CB, Angelini A, Berry GJ, Burke MM, Demetris AJ, Hammond E, Itescu S, Marboe CC, McManus B, Reed EF, Reinsmoen NL, Rodriguez ER, Rose AG, Rose M, Suciu-Focia N, Zeevi A and Billingham ME. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–20. - PubMed
-
- Berry GJ, Burke MM, Andersen C, Bruneval P, Fedrigo M, Fishbein MC, Goddard M, Hammond EH, Leone O, Marboe C, Miller D, Neil D, Rassl D, Revelo MP, Rice A, Rene Rodriguez E, Stewart S, Tan CD, Winters GL, West L, Mehra MR and Angelini A. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Heart Lung Transplant. 2013;32:1147–62. - PubMed
-
- Marboe CC, Billingham M, Eisen H, Deng MC, Baron H, Mehra M, Hunt S, Wohlgemuth J, Mahmood I, Prentice J and Berry G. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24:S219–26. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

