The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases
- PMID: 33435894
- PMCID: PMC7805029
- DOI: 10.1186/s12879-020-05746-5
The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases
Abstract
Background: Although traditional diagnostic techniques of infection are mature and price favorable at present, most of them are time-consuming and with a low positivity. Metagenomic next⁃generation sequencing (mNGS) was studied widely because of identification and typing of all pathogens not rely on culture and retrieving all DNA without bias. Based on this background, we aim to detect the difference between mNGS and traditional culture method, and to explore the relationship between mNGS results and the severity, prognosis of infectious patients.
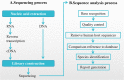
Methods: 109 adult patients were enrolled in our study in Shanghai Tenth People's Hospital from October 2018 to December 2019. The diagnostic results, negative predictive values, positive predictive values, false positive rate, false negative rate, pathogen and sample types were analyzed by using both traditional culture and mNGS methods. Then, the samples and clinical information of 93 patients in the infected group (ID) were collected. According to whether mNGS detected pathogens, the patients in ID group were divided into the positive group of 67 cases and the negative group of 26 cases. Peripheral blood leukocytes, C-reactive protein (CRP), procalcitonin (PCT) and neutrophil counts were measured, and the concentrations of IL-2, IL-4, IL-6, TNF-α, IL-17A, IL-10 and INF-γ in the serum were determined by ELISA. The correlation between the positive detection of pathogens by mNGS and the severity of illness, hospitalization days, and mortality were analyzed.
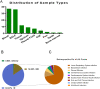
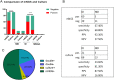
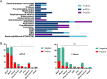
Results: 109 samples were assigned into infected group (ID, 92/109, 84.4%), non-infected group (NID, 16/109, 14.7%), and unknown group (1/109, 0.9%). Blood was the most abundant type of samples with 37 cases, followed by bronchoalveolar lavage fluid in 36 cases, tissue, sputum, pleural effusion, cerebrospinal fluid (CSF), pus, bone marrow and nasal swab. In the ID group, the majority of patients were diagnosed with lower respiratory system infections (73/109, 67%), followed by bloodstream infections, pleural effusion and central nervous system infections. The sensitivity of mNGS was significantly higher than that of culture method (67.4% vs 23.6%; P < 0.001), especially in sample types of bronchoalveolar lavage fluid (P = 0.002), blood (P < 0.001) and sputum (P = 0.037), while the specificity of mNGS was not significantly different from culture method (68.8% vs 81.3%; P = 0.41). The number of hospitals stays and 28-day-motality in the positive mNGS group were significantly higher than those in the negative group, and the difference was statistically significant (P < 0.05). Age was significant in multivariate logistic analyses of positive results of mNGS.
Conclusions: The study found that mNGS had a higher sensitivity than the traditional method, especially in blood, bronchoalveolar lavage fluid and sputum samples. And positive mNGS group had a higher hospital stay, 28-day-mortality, which means the positive of pathogen nucleic acid sequences detection may be a potential high-risk factor for poor prognosis of adult patients and has significant clinical value. MNGS should be used more in early pathogen diagnosis in the future.
Keywords: Diagnostic; Infection; Next-generation sequencing; Sensitivity; Survival.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, Raangs EG, Lo-Ten-Foe J, Grundmann H, Rossen JW, Friedrich AW. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum beta-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep. 2016;6:20840. doi: 10.1038/srep20840. - DOI - PMC - PubMed
-
- Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice C, Committee on Laboratory Practices of the American Society for M, Microbiology Resource Committee of the College of American P. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 2017; 141:776–786. - PubMed
-
- Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical Practice. Clin Infect Dis. 2018;67:S231–S240. doi: 10.1093/cid/ciy693. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

