Integration of sperm ncRNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance
- PMID: 33436057
- PMCID: PMC7802319
- DOI: 10.1186/s13072-020-00378-0
Integration of sperm ncRNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance
Abstract
Background: Environmentally induced epigenetic transgenerational inheritance of pathology and phenotypic variation has been demonstrated in all organisms investigated from plants to humans. This non-genetic form of inheritance is mediated through epigenetic alterations in the sperm and/or egg to subsequent generations. Although the combined regulation of differential DNA methylated regions (DMR), non-coding RNA (ncRNA), and differential histone retention (DHR) have been shown to occur, the integration of these different epigenetic processes remains to be elucidated. The current study was designed to examine the integration of the different epigenetic processes.
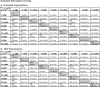
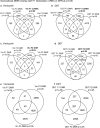
Results: A rat model of transiently exposed F0 generation gestating females to the agricultural fungicide vinclozolin or pesticide DDT (dichloro-diphenyl-trichloroethane) was used to acquire the sperm from adult males in the subsequent F1 generation offspring, F2 generation grand offspring, and F3 generation great-grand offspring. The F1 generation sperm ncRNA had substantial overlap with the F1, F2 and F3 generation DMRs, suggesting a potential role for RNA-directed DNA methylation. The DMRs also had significant overlap with the DHRs, suggesting potential DNA methylation-directed histone retention. In addition, a high percentage of DMRs induced in the F1 generation sperm were maintained in subsequent generations.
Conclusions: Many of the DMRs, ncRNA, and DHRs were colocalized to the same chromosomal location regions. Observations suggest an integration of DMRs, ncRNA, and DHRs in part involve RNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance.
Keywords: DNA methylation; Epigenetics; Histone; Non-genetic inheritance; Review; Sperm; Transgenerational; ncRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

