Post-transcriptional circadian regulation in macrophages organizes temporally distinct immunometabolic states
- PMID: 33436377
- PMCID: PMC7849412
- DOI: 10.1101/gr.263814.120
Post-transcriptional circadian regulation in macrophages organizes temporally distinct immunometabolic states
Abstract
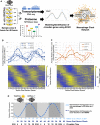
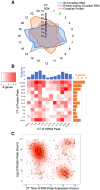
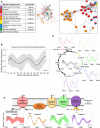
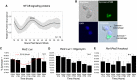
Our core timekeeping mechanism, the circadian clock, plays a vital role in immunity. Although the mechanics of circadian control over the immune response is generally explained by transcriptional activation or repression derived from this clock's transcription-translation negative-feedback loop, research suggests that some regulation occurs beyond transcriptional activity. We comprehensively profiled the transcriptome and proteome of murine bone marrow-derived macrophages and found that only 15% of the circadian proteome had corresponding oscillating mRNA, suggesting post-transcriptional regulation influences macrophage clock regulatory output to a greater extent than any other tissue previously profiled. This regulation may be explained by the robust temporal enrichment we identified for proteins involved in degradation and translation. Extensive post-transcriptional temporal-gating of metabolic pathways was also observed and further corresponded with daily variations in ATP production, mitochondrial morphology, and phagocytosis. The disruption of this circadian post-transcriptional metabolic regulation impaired immune functionality. Our results demonstrate that cell-intrinsic post-transcriptional regulation is a primary driver of circadian output in macrophages and that this regulation, particularly of metabolic pathways, plays an important role in determining their response to immune stimuli.
© 2021 Collins et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
Cited by
-
Circadian control of heparan sulfate levels times phagocytosis of amyloid beta aggregates.PLoS Genet. 2022 Feb 10;18(2):e1009994. doi: 10.1371/journal.pgen.1009994. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35143487 Free PMC article.
-
The Circadian Clock Protein BMAL1 Acts as a Metabolic Sensor In Macrophages to Control the Production of Pro IL-1β.Front Immunol. 2021 Nov 9;12:700431. doi: 10.3389/fimmu.2021.700431. eCollection 2021. Front Immunol. 2021. PMID: 34858390 Free PMC article.
-
The circadian clock influences T cell responses to vaccination by regulating dendritic cell antigen processing.Nat Commun. 2022 Dec 5;13(1):7217. doi: 10.1038/s41467-022-34897-z. Nat Commun. 2022. PMID: 36470865 Free PMC article.
-
Circadian rhythms in adaptive immunity and vaccination.Semin Immunopathol. 2022 Mar;44(2):193-207. doi: 10.1007/s00281-021-00903-7. Epub 2021 Nov 25. Semin Immunopathol. 2022. PMID: 34825270 Review.
-
Macrophage circadian rhythms are differentially affected based on stimuli.Integr Biol (Camb). 2022 Jun 8;14(3):62-75. doi: 10.1093/intbio/zyac007. Integr Biol (Camb). 2022. PMID: 35652485 Free PMC article.
References
-
- Abbas A, Lichtman A, Pillai, S. 2018. Innate immunity. In Cellular and molecular immunology, 9th ed (ed. Merritt J), pp. 82–90. Elsevier, Philadelphia.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
