Dinoroseobacter shibae Outer Membrane Vesicles Are Enriched for the Chromosome Dimer Resolution Site dif
- PMID: 33436507
- PMCID: PMC7901474
- DOI: 10.1128/mSystems.00693-20
Dinoroseobacter shibae Outer Membrane Vesicles Are Enriched for the Chromosome Dimer Resolution Site dif
Abstract
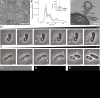
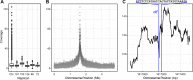
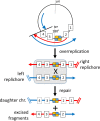
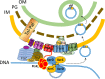
Outer membrane vesicles (OMVs) are universally produced by prokaryotes and play important roles in symbiotic and pathogenic interactions. They often contain DNA, but a mechanism for its incorporation is lacking. Here, we show that Dinoroseobacter shibae, a dinoflagellate symbiont, constitutively secretes OMVs containing DNA. Time-lapse microscopy captured instances of multiple OMV production at the septum during cell division. DNA from the vesicle lumen was up to 22-fold enriched for the region around the terminus of replication (ter). The peak of coverage was located at dif, a conserved 28-bp palindromic sequence required for binding of the site-specific tyrosine recombinases XerC/XerD. These enzymes are activated at the last stage of cell division immediately prior to septum formation when they are bound by the divisome protein FtsK. We suggest that overreplicated regions around the terminus have been repaired by the FtsK-dif-XerC/XerD molecular machinery. The vesicle proteome was clearly dominated by outer membrane and periplasmic proteins. Some of the most abundant vesicle membrane proteins were predicted to be required for direct interaction with peptidoglycan during cell division (LysM, Tol-Pal, Spol, lytic murein transglycosylase). OMVs were 15-fold enriched for the saturated fatty acid 16:00. We hypothesize that constitutive OMV secretion in D. shibae is coupled to cell division. The footprint of the FtsK-dif-XerC/XerD molecular machinery suggests a novel potentially highly conserved route for incorporation of DNA into OMVs. Clearing the division site from small DNA fragments might be an important function of vesicles produced during exponential growth under optimal conditions.IMPORTANCE Gram-negative bacteria continually form vesicles from their outer membrane (outer membrane vesicles [OMVs]) during normal growth. OMVs frequently contain DNA, and it is unclear how DNA can be shuffled from the cytoplasm to the OMVs. We studied OMV cargo in Dinoroseobacter shibae, a symbiont of dinoflagellates, using microscopy and a multi-omics approach. We found that vesicles formed during undisturbed exponential growth contain DNA which is enriched for genes around the replication terminus, specifically, the binding site for an enzyme complex that is activated at the last stage of cell division. We suggest that the enriched genes are the result of overreplication which is repaired by their excision and excretion via membrane vesicles to clear the divisome from waste DNA.
Keywords: DNA repair; OMV; circular chromosomes; replication termination; vesicles.
Copyright © 2021 Wang et al.
Figures

Similar articles
-
The effect of site-specific recombinases XerCD on the removal of over-replicated chromosomal DNA through outer membrane vesicles in bacteria.Microbiol Spectr. 2024 Mar 5;12(3):e0234323. doi: 10.1128/spectrum.02343-23. Epub 2024 Feb 13. Microbiol Spectr. 2024. PMID: 38349173 Free PMC article.
-
Multilamellar and Multivesicular Outer Membrane Vesicles Produced by a Buttiauxella agrestis tolB Mutant.Appl Environ Microbiol. 2020 Oct 1;86(20):e01131-20. doi: 10.1128/AEM.01131-20. Print 2020 Oct 1. Appl Environ Microbiol. 2020. PMID: 32801184 Free PMC article.
-
Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli.J Mol Biol. 1997 Feb 28;266(3):525-37. doi: 10.1006/jmbi.1996.0828. J Mol Biol. 1997. PMID: 9067608
-
Xer Site Specific Recombination: Double and Single Recombinase Systems.Front Microbiol. 2017 Mar 20;8:453. doi: 10.3389/fmicb.2017.00453. eCollection 2017. Front Microbiol. 2017. PMID: 28373867 Free PMC article. Review.
-
Protein selection and export via outer membrane vesicles.Biochim Biophys Acta. 2014 Aug;1843(8):1612-9. doi: 10.1016/j.bbamcr.2013.12.011. Epub 2013 Dec 24. Biochim Biophys Acta. 2014. PMID: 24370777 Free PMC article. Review.
Cited by
-
Unravelling the DNA sequences carried by Streptomyces coelicolor membrane vesicles.Sci Rep. 2022 Oct 5;12(1):16651. doi: 10.1038/s41598-022-21002-z. Sci Rep. 2022. PMID: 36198712 Free PMC article.
-
The effect of site-specific recombinases XerCD on the removal of over-replicated chromosomal DNA through outer membrane vesicles in bacteria.Microbiol Spectr. 2024 Mar 5;12(3):e0234323. doi: 10.1128/spectrum.02343-23. Epub 2024 Feb 13. Microbiol Spectr. 2024. PMID: 38349173 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
