Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes
- PMID: 33436567
- PMCID: PMC7804181
- DOI: 10.1038/s41467-020-20237-6
Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes
Erratum in
-
Publisher Correction: Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes.Nat Commun. 2021 Feb 8;12(1):988. doi: 10.1038/s41467-021-21294-1. Nat Commun. 2021. PMID: 33558551 Free PMC article. No abstract available.
Abstract
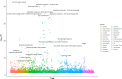
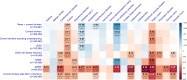
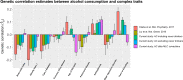
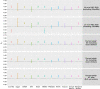
Genome-wide association studies (GWAS) have discovered numerous genetic variants associated with human behavioural traits. However, behavioural traits are subject to misreports and longitudinal changes (MLC) which can cause biases in GWAS and follow-up analyses. Here, we demonstrate that individuals with higher disease burden in the UK Biobank (n = 455,607) are more likely to misreport or reduce their alcohol consumption levels, and propose a correction procedure to mitigate the MLC-induced biases. The alcohol consumption GWAS signals removed by the MLC corrections are enriched in metabolic/cardiovascular traits. Almost all the previously reported negative estimates of genetic correlations between alcohol consumption and common diseases become positive/non-significant after the MLC corrections. We also observe MLC biases for smoking and physical activities in the UK Biobank. Our findings provide a plausible explanation of the controversy about the effects of alcohol consumption on health outcomes and a caution for future analyses of self-reported behavioural traits in biobank data.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Burton R, Sheron N. No level of alcohol consumption improves health. Lancet. 2018;392:987–988. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

