Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis
- PMID: 33436578
- PMCID: PMC7804935
- DOI: 10.1038/s41467-020-20565-7
Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis
Abstract
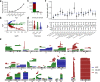
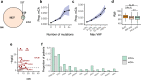
Stably acquired mutations in hematopoietic cells represent substrates of selection that may lead to clonal hematopoiesis (CH), a common state in cancer patients that is associated with a heightened risk of leukemia development. Owing to technical and sample size limitations, most CH studies have characterized gene mutations or mosaic chromosomal alterations (mCAs) individually. Here we leverage peripheral blood sequencing data from 32,442 cancer patients to jointly characterize gene mutations (n = 14,789) and mCAs (n = 383) in CH. Recurrent composite genotypes resembling known genetic interactions in leukemia genomes underlie 23% of all detected autosomal alterations, indicating that these selection mechanisms are operative early in clonal evolution. CH with composite genotypes defines a patient group at high risk of leukemia progression (3-year cumulative incidence 14.6%, CI: 7-22%). Multivariable analysis identifies mCA as an independent risk factor for leukemia development (HR = 14, 95% CI: 6-33, P < 0.001). Our results suggest that mCA should be considered in conjunction with gene mutations in the surveillance of patients at risk of hematologic neoplasms.
Conflict of interest statement
The authors declare the following competing interests: K.L.B. has received research funding from GRAIL; E.B. receives research funding from Celgene. D.B.S. has served as a consultant/received honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Illumina, and Vivideon Therapeutics. M.F.B has participated in advisory board activities for Roche and has received research support from Grail and Illumina. S.M.D. is principal owner of Daboia Consulting LLC. L.A.D. is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics. He is a paid consultant to PGDx and Neophore. He is an uncompensated consultant for Merck but has received travel and research support for clinical trials from Merck. L.A.D. is an inventor of multiple licensed patents related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy from Johns Hopkins University. Some of these licenses and relationships are associated with equity or royalty payments directly to Johns Hopkins and L.A.D. His wife holds equity in Amgen. The terms of all these arrangements are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict of interest policies. J.S.M.M. is a member of the board of directors and holds equity in Isabl, a software analytics company for high-throughput clinical whole-genome and RNA-sequencing analyses. R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics, and Isoplexis which include equity interest. He receives research support from and consulted for Celgene and Roche, and has consulted for Lilly, Janssen, Astellas, Morphosys, and Novartis. He has received honoraria from Roche, Lilly, and Amgen for invited lectures and from Gilead for grant reviews. A.Z. received honoraria from Illumina. E. Papaemmanuil receives research funding from Celgene and has received honoraria for speaking and scientific advisory engagements with Celgene, Prime Oncology, Novartis, Illumina, and Kyowa Hakko Kirin. E. Papaemmanuil is also a member of the board of directors and holds equity in Isabl, a software analytics company for high-throughput clinical whole-genome and RNA-sequencing analyses. E. Papaemmanuil is an inventor in software licenses related to technology of genome analytics. Some of these licenses and relationships are associated with equity or royalty payments to MSKCC and are managed accordingly by the conflict of interest office at MSKCC. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

