isoCirc catalogs full-length circular RNA isoforms in human transcriptomes
- PMID: 33436621
- PMCID: PMC7803736
- DOI: 10.1038/s41467-020-20459-8
isoCirc catalogs full-length circular RNA isoforms in human transcriptomes
Abstract
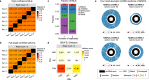
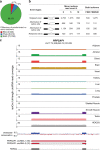
Circular RNAs (circRNAs) have emerged as an important class of functional RNA molecules. Short-read RNA sequencing (RNA-seq) is a widely used strategy to identify circRNAs. However, an inherent limitation of short-read RNA-seq is that it does not experimentally determine the full-length sequences and exact exonic compositions of circRNAs. Here, we report isoCirc, a strategy for sequencing full-length circRNA isoforms, using rolling circle amplification followed by nanopore long-read sequencing. We describe an integrated computational pipeline to reliably characterize full-length circRNA isoforms using isoCirc data. Using isoCirc, we generate a comprehensive catalog of 107,147 full-length circRNA isoforms across 12 human tissues and one human cell line (HEK293), including 40,628 isoforms ≥500 nt in length. We identify widespread alternative splicing events within the internal part of circRNAs, including 720 retained intron events corresponding to a class of exon-intron circRNAs (EIciRNAs). Collectively, isoCirc and the companion dataset provide a useful strategy and resource for studying circRNAs in human transcriptomes.
Conflict of interest statement
Y.X. is a scientific cofounder of Panorama Medicine.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

