Outer membrane vesicles containing OmpA induce mitochondrial fragmentation to promote pathogenesis of Acinetobacter baumannii
- PMID: 33436835
- PMCID: PMC7804284
- DOI: 10.1038/s41598-020-79966-9
Outer membrane vesicles containing OmpA induce mitochondrial fragmentation to promote pathogenesis of Acinetobacter baumannii
Abstract
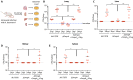
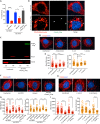
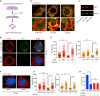
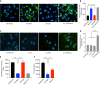
Acinetobacter baumannii is a highly antibiotic resistant Gram-negative bacterium that causes life-threatening infections in humans with a very high mortality rate. A. baumannii is an extracellular pathogen with poorly understood virulence mechanisms. Here we report that A. baumannii employs the release of outer membrane vesicles (OMVs) containing the outer membrane protein A (OmpAAb) to promote bacterial pathogenesis and dissemination. OMVs containing OmpAAb are taken up by mammalian cells where they activate the host GTPase dynamin-related protein 1 (DRP1). OmpAAb mediated activation of DRP1 enhances its accumulation on mitochondria that causes mitochondrial fragmentation, elevation in reactive oxygen species (ROS) production and cell death. Loss of DRP1 rescues these phenotypes. Our data show that OmpAAb is sufficient to induce mitochondrial fragmentation and cytotoxicity since its expression in E. coli transfers its pathogenic properties to E. coli. A. baumannii infection in mice also induces mitochondrial damage in alveolar macrophages in an OmpAAb dependent manner. We finally show that OmpAAb is also required for systemic dissemination in the mouse lung infection model. In this study we uncover the mechanism of OmpAAb as a virulence factor in A. baumannii infections and further establish the host cell factor required for its pathogenic effects.
Conflict of interest statement
The authors declare no competing interests.
Figures
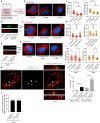
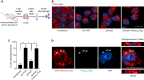
Similar articles
-
Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles.PLoS One. 2011 Feb 28;6(2):e17027. doi: 10.1371/journal.pone.0017027. PLoS One. 2011. PMID: 21386968 Free PMC article.
-
Acinetobacter baumannii OmpA Is a Selective Antibiotic Permeant Porin.ACS Infect Dis. 2018 Mar 9;4(3):373-381. doi: 10.1021/acsinfecdis.7b00168. Epub 2017 Dec 26. ACS Infect Dis. 2018. PMID: 29260856
-
Thioredoxin-mediated alteration of protein content and cytotoxicity of Acinetobacter baumannii outer membrane vesicles.Exp Biol Med (Maywood). 2022 Feb;247(3):282-288. doi: 10.1177/15353702211052952. Epub 2021 Oct 29. Exp Biol Med (Maywood). 2022. PMID: 34713732 Free PMC article.
-
Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection.J Biomed Sci. 2020 Jan 18;27(1):26. doi: 10.1186/s12929-020-0617-7. J Biomed Sci. 2020. PMID: 31954394 Free PMC article. Review.
-
Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models.FEMS Microbiol Rev. 2013 Mar;37(2):130-55. doi: 10.1111/j.1574-6976.2012.00344.x. Epub 2012 Jun 18. FEMS Microbiol Rev. 2013. PMID: 22568581 Review.
Cited by
-
Co-regulation of biofilm formation and antimicrobial resistance in Acinetobacter baumannii: from mechanisms to therapeutic strategies.Eur J Clin Microbiol Infect Dis. 2023 Dec;42(12):1405-1423. doi: 10.1007/s10096-023-04677-8. Epub 2023 Oct 28. Eur J Clin Microbiol Infect Dis. 2023. PMID: 37897520 Free PMC article. Review.
-
Bacterial membrane vesicles: orchestrators of interkingdom interactions in microbial communities for environmental adaptation and pathogenic dynamics.Front Immunol. 2024 Mar 21;15:1371317. doi: 10.3389/fimmu.2024.1371317. eCollection 2024. Front Immunol. 2024. PMID: 38576623 Free PMC article. Review.
-
Emerging role of bacterial outer membrane vesicle in gastrointestinal tract.Gut Pathog. 2023 Apr 27;15(1):20. doi: 10.1186/s13099-023-00543-2. Gut Pathog. 2023. PMID: 37106359 Free PMC article. Review.
-
Outer Membrane Vesicles from Acinetobacter baumannii: Biogenesis, Functions, and Vaccine Application.Vaccines (Basel). 2023 Dec 31;12(1):49. doi: 10.3390/vaccines12010049. Vaccines (Basel). 2023. PMID: 38250862 Free PMC article. Review.
-
Association between the Respiratory Microbiome and Plasma Microbial Extracellular Vesicles in Intubated Patients.Microorganisms. 2023 Aug 22;11(9):2128. doi: 10.3390/microorganisms11092128. Microorganisms. 2023. PMID: 37763972 Free PMC article.
References
-
- Tacconelli E, Margrini N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO. 2013 doi: 10.1590/S0100-15742013000100018. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

