The N-terminal region of Jaw1 has a role to inhibit the formation of organized smooth endoplasmic reticulum as an intrinsically disordered region
- PMID: 33436890
- PMCID: PMC7804115
- DOI: 10.1038/s41598-020-80258-5
The N-terminal region of Jaw1 has a role to inhibit the formation of organized smooth endoplasmic reticulum as an intrinsically disordered region
Abstract
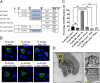
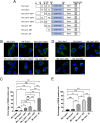
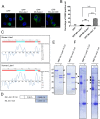
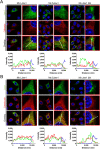
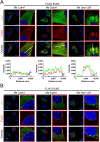
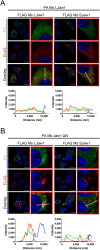
Jaw1/LRMP is a type II integral membrane protein that is localized at the endoplasmic reticulum (ER) and outer nuclear membrane. We previously reported that a function of Jaw1 is to maintain the nuclear shape as a KASH protein via its carboxyl terminal region, a component of linker of nucleoskeleton and cytoskeleton complex in the oligomeric state. Although the oligomerization of some KASH proteins via the cytosolic regions serves to stabilize protein-protein interactions, the issue of how the oligomerization of Jaw1 is regulated is not completely understood. Therefore, we focused on three distinct regions on the cytosolic face of Jaw1: the N-terminal region, the coiled-coil domain and the stem region, in terms of oligomerization. A co-immunoprecipitation assay showed that its coiled-coil domain is a candidate for the oligomerization site. Furthermore, our data indicated that the N-terminal region prevents the aberrant oligomerization of Jaw1 as an intrinsically disordered region (IDR). Importantly, the ectopic expression of an N-terminal region deleted mutant caused the formation of organized smooth ER (OSER), structures such as nuclear karmellae and whorls, in B16F10 cells. Furthermore, this OSER interfered with the localization of the oligomer and interactors such as the type III inositol 1,4,5-triphosphate receptor (IP3R3) and SUN2. In summary, the N-terminal region of Jaw1 inhibits the formation of OSER as an IDR to maintain the homeostatic localization of interactors on the ER membrane.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Jaw1/LRMP has a role in maintaining nuclear shape via interaction with SUN proteins.J Biochem. 2018 Oct 1;164(4):303-311. doi: 10.1093/jb/mvy053. J Biochem. 2018. PMID: 29878215
-
Jaw1/LRMP is associated with the maintenance of Golgi ribbon structure.J Biochem. 2023 Apr 26;173(5):383-392. doi: 10.1093/jb/mvad004. J Biochem. 2023. PMID: 36689741
-
Cleavage of the Jaw1 C-terminal region enhances its augmentative effect on the Ca2+ release via IP3 receptors.J Cell Sci. 2023 Feb 15;136(4):jcs260439. doi: 10.1242/jcs.260439. Epub 2023 Feb 15. J Cell Sci. 2023. PMID: 36789796
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
-
The disordered boundary of the cell: emerging properties of membrane-bound intrinsically disordered proteins.Biomol Concepts. 2019 Apr 3;10(1):25-36. doi: 10.1515/bmc-2019-0003. Biomol Concepts. 2019. PMID: 30956226 Review.
Cited by
-
Novel Functional Features of cGMP Substrate Proteins IRAG1 and IRAG2.Int J Mol Sci. 2023 Jun 7;24(12):9837. doi: 10.3390/ijms24129837. Int J Mol Sci. 2023. PMID: 37372987 Free PMC article. Review.
-
Jaw1 accelerates the reaction speed of the Ca2+ signals via ITPRs upon GPCR stimulation.Sci Rep. 2025 Mar 24;15(1):10104. doi: 10.1038/s41598-025-94489-x. Sci Rep. 2025. PMID: 40128249 Free PMC article.
-
Jaw1/LRMP increases Ca2+ influx upon GPCR stimulation with heterogeneous effect on the activity of each ITPR subtype.Sci Rep. 2022 Jun 8;12(1):9476. doi: 10.1038/s41598-022-13620-4. Sci Rep. 2022. PMID: 35676525 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

