A deep learning framework combined with word embedding to identify DNA replication origins
- PMID: 33436981
- PMCID: PMC7804333
- DOI: 10.1038/s41598-020-80670-x
A deep learning framework combined with word embedding to identify DNA replication origins
Abstract
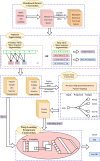
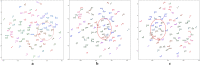
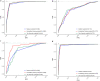
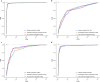
The DNA replication influences the inheritance of genetic information in the DNA life cycle. As the distribution of replication origins (ORIs) is the major determinant to precisely regulate the replication process, the correct identification of ORIs is significant in giving an insightful understanding of DNA replication mechanisms and the regulatory mechanisms of genetic expressions. For eukaryotes in particular, multiple ORIs exist in each of their gene sequences to complete the replication in a reasonable period of time. To simplify the identification process of eukaryote's ORIs, most of existing methods are developed by traditional machine learning algorithms, and target to the gene sequences with a fixed length. Consequently, the identification results are not satisfying, i.e. there is still great room for improvement. To break through the limitations in previous studies, this paper develops sequence segmentation methods, and employs the word embedding technique, 'Word2vec', to convert gene sequences into word vectors, thereby grasping the inner correlations of gene sequences with different lengths. Then, a deep learning framework to perform the ORI identification task is constructed by a convolutional neural network with an embedding layer. On the basis of the analysis of similarity reduction dimensionality diagram, Word2vec can effectively transform the inner relationship among words into numerical feature. For four species in this study, the best models are obtained with the overall accuracy of 0.975, 0.765, 0.885, 0.967, the Matthew's correlation coefficient of 0.940, 0.530, 0.771, 0.934, and the AUC of 0.975, 0.800, 0.888, 0.981, which indicate that the proposed predictor has a stable ability and provide a high confidence coefficient to classify both of ORIs and non-ORIs. Compared with state-of-the-art methods, the proposed predictor can achieve ORI identification with significant improvement. It is therefore reasonable to anticipate that the proposed method will make a useful high throughput tool for genome analysis.
Conflict of interest statement
The authors declare no competing interests.
Figures



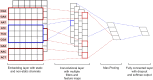


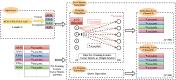

Similar articles
-
iEnhancer-GAN: A Deep Learning Framework in Combination with Word Embedding and Sequence Generative Adversarial Net to Identify Enhancers and Their Strength.Int J Mol Sci. 2021 Mar 30;22(7):3589. doi: 10.3390/ijms22073589. Int J Mol Sci. 2021. PMID: 33808317 Free PMC article.
-
A deep learning framework for enhancer prediction using word embedding and sequence generation.Biophys Chem. 2022 Jul;286:106822. doi: 10.1016/j.bpc.2022.106822. Epub 2022 May 5. Biophys Chem. 2022. PMID: 35605495
-
Origin replication complex binding, nucleosome depletion patterns, and a primary sequence motif can predict origins of replication in a genome with epigenetic centromeres.mBio. 2014 Sep 2;5(5):e01703-14. doi: 10.1128/mBio.01703-14. mBio. 2014. PMID: 25182328 Free PMC article.
-
Genomic specification and epigenetic regulation of eukaryotic DNA replication origins.EMBO J. 2004 Nov 10;23(22):4365-70. doi: 10.1038/sj.emboj.7600450. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510221 Free PMC article. Review.
-
Recent Advances on the Machine Learning Methods in Identifying DNA Replication Origins in Eukaryotic Genomics.Front Genet. 2018 Dec 10;9:613. doi: 10.3389/fgene.2018.00613. eCollection 2018. Front Genet. 2018. PMID: 30619452 Free PMC article. Review.
Cited by
-
DNA sequence analysis landscape: a comprehensive review of DNA sequence analysis task types, databases, datasets, word embedding methods, and language models.Front Med (Lausanne). 2025 Apr 8;12:1503229. doi: 10.3389/fmed.2025.1503229. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40265190 Free PMC article. Review.
-
Deep learning and support vector machines for transcription start site identification.PeerJ Comput Sci. 2023 Apr 17;9:e1340. doi: 10.7717/peerj-cs.1340. eCollection 2023. PeerJ Comput Sci. 2023. PMID: 37346545 Free PMC article.
-
Genomic Surveillance of COVID-19 Variants With Language Models and Machine Learning.Front Genet. 2022 Apr 8;13:858252. doi: 10.3389/fgene.2022.858252. eCollection 2022. Front Genet. 2022. PMID: 35464852 Free PMC article.
-
Investigation of cell development and tissue structure network based on natural Language processing of scRNA-seq data.J Transl Med. 2025 Mar 4;23(1):264. doi: 10.1186/s12967-025-06263-2. J Transl Med. 2025. PMID: 40038714 Free PMC article.
-
Discovering genomic islands in unannotated bacterial genomes using sequence embedding.Bioinform Adv. 2024 Jun 17;4(1):vbae089. doi: 10.1093/bioadv/vbae089. eCollection 2024. Bioinform Adv. 2024. PMID: 38911822 Free PMC article.
References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

