Identification of circ-FAM169A sponges miR-583 involved in the regulation of intervertebral disc degeneration
- PMID: 33437631
- PMCID: PMC7773979
- DOI: 10.1016/j.jot.2020.07.007
Identification of circ-FAM169A sponges miR-583 involved in the regulation of intervertebral disc degeneration
Abstract
Objective: Low back pain (LBP) is the predominant cause of disc degeneration in patients, which brings serious social problems and economic burdens. Increasing evidence has indicated that intervertebral disc degeneration (IDD) is one of the most common causes triggering LBP. Accumulating evidence has shown that circRNAs are involved in the pathological process of IDD. Nevertheless, the circRNA-mediated IDD pathogenesis still remains unknown. This study explored the potential mechanism and functions of circ-FAM169A in NPCs.
Methods: Bioinformatics analysis was conducted to identify key circRNA, miRNA and mRNA and predict their potential role in IDD. Dual-luciferase reporter assay, western blot, qRT-PCR, and fluorescence in situ hybridisation (FISH) were used to demonstrate the interaction among circ-FAM169A, miR-583 and Sox9 in NPCs.
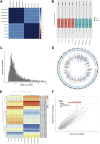
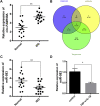
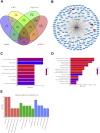
Results: Herein, we identified circ-FAM169A, which was dramatically up-regulated in degenerative nucleus pulposus (NP) tissues and negatively correlated with expression levels of miR-583. We constructed a circ-FAM169A-miR-583-mRNAs co-expression network and predicted circ-FAM169A-miR-583 pathway predominantly involved in extracellular matrix metabolism and cell apoptosis etc. FISH experiments confirmed circ-FAM169A and miR-583 co-existence in the cytoplasm of NPCs. Luciferase reporter assay illustrated that circ-FAM169A was directly bound to miR-583 and Sox9 was the directly target gene of miR-583. Additionally, miR-583 negatively regulated Sox9 mRNA and protein levels in NPCs.
Conclusion: Findings of this study indicated that circ-FAM169A-miR-583 pathway may play a significant role in the regulation of IDD, which will provide novel diagnostic biomarkers and develop effective treatment strategy of IDD diseases.
The translational potential of this article: This study suggested that circ-FAM169A-miR-583 pathway may regulate NPCs apoptosis and extracellular matrix synthesis and catabolism by targeting Sox9. It provides a novel therapeutic target and strategy for IVDD diseases.
Keywords: Competitive endogenous RNA; Differential expressed circRNAs; GEO; Intervertebral disc degeneration; circ-FAM169A; miR-583.
© 2020 The Authors.
Conflict of interest statement
The authors have no conflicts of interest to disclose in relation to this article.
Figures




Similar articles
-
The circular RNA FAM169A functions as a competitive endogenous RNA and regulates intervertebral disc degeneration by targeting miR-583 and BTRC.Cell Death Dis. 2020 May 4;11(5):315. doi: 10.1038/s41419-020-2543-8. Cell Death Dis. 2020. PMID: 32366862 Free PMC article.
-
Hsa_circ_0001946 Ameliorates Mechanical Stress-induced Intervertebral Disk Degeneration Via Targeting miR-432-5p and SOX9.Spine (Phila Pa 1976). 2023 Dec 1;48(23):E401-E408. doi: 10.1097/BRS.0000000000004777. Epub 2023 Aug 9. Spine (Phila Pa 1976). 2023. PMID: 37555796 Free PMC article.
-
circ_0000389 inhibits intervertebral disc degeneration by targeting the miR-346/KLF7 axis.Acta Biochim Biophys Sin (Shanghai). 2025 Mar 7. doi: 10.3724/abbs.2025029. Online ahead of print. Acta Biochim Biophys Sin (Shanghai). 2025. PMID: 40055913
-
The pathological mechanisms of circRNAs in mediating intervertebral disc degeneration.Noncoding RNA Res. 2023 Sep 18;8(4):633-640. doi: 10.1016/j.ncrna.2023.09.004. eCollection 2023 Dec. Noncoding RNA Res. 2023. PMID: 37780894 Free PMC article. Review.
-
Emerging evidence on noncoding-RNA regulatory machinery in intervertebral disc degeneration: a narrative review.Arthritis Res Ther. 2020 Nov 16;22(1):270. doi: 10.1186/s13075-020-02353-2. Arthritis Res Ther. 2020. PMID: 33198793 Free PMC article. Review.
Cited by
-
Sinuvertebral nerve block treats discogenic low back pain: a retrospective cohort study.Ann Transl Med. 2022 Nov;10(22):1219. doi: 10.21037/atm-22-5297. Ann Transl Med. 2022. PMID: 36544669 Free PMC article.
-
CircZNF418 alleviates oxidative stress-induced cartilage endplate degeneration by stabilizing Sox9 through HuR.J Mol Histol. 2025 Jul 31;56(4):242. doi: 10.1007/s10735-025-10528-x. J Mol Histol. 2025. PMID: 40742571
-
Regulated cell death: Implications for intervertebral disc degeneration and therapy.J Orthop Translat. 2022 Nov 5;37:163-172. doi: 10.1016/j.jot.2022.10.009. eCollection 2022 Nov. J Orthop Translat. 2022. PMID: 36380883 Free PMC article. Review.
-
Identification of Differentially Expressed circRNAs, miRNAs, and Genes in Patients Associated with Cartilaginous Endplate Degeneration.Biomed Res Int. 2021 May 18;2021:2545459. doi: 10.1155/2021/2545459. eCollection 2021. Biomed Res Int. 2021. PMID: 34104646 Free PMC article.
-
Role of the miR-133a-5p/FBXO6 axis in the regulation of intervertebral disc degeneration.J Orthop Translat. 2021 Jun 19;29:123-133. doi: 10.1016/j.jot.2021.05.004. eCollection 2021 Jul. J Orthop Translat. 2021. PMID: 34249610 Free PMC article.
References
-
- James S.L., Abate D., Abate K.H., Abay S.M., Abbafati C., Abbasi N. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. - PMC - PubMed
-
- Andersson Gunnar B.J. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. - PubMed
-
- Martin B.I., Deyo R.A., Mirza S.K., Turner J.A., Mirza S.K., Comstock B.A. Expenditures and health status among adults with back and neck problems. J Am Med Assoc. 2008;299:656–664. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

