Enfuvirtide, an HIV-1 fusion inhibitor peptide, can act as a potent SARS-CoV-2 fusion inhibitor: an in silico drug repurposing study
- PMID: 33438525
- PMCID: PMC7814568
- DOI: 10.1080/07391102.2021.1871958
Enfuvirtide, an HIV-1 fusion inhibitor peptide, can act as a potent SARS-CoV-2 fusion inhibitor: an in silico drug repurposing study
Abstract
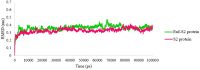
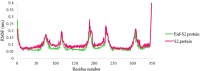
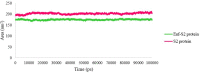
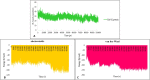
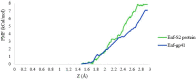
Regarding the urgency of therapeutic measures for coronavirus disease 2019 (COVID-19) pandemic, the use of available drugs with FDA approval is preferred because of the less time and cost required for their development. In silico drug repurposing is an accurate way to speed up the screening of the existing FDA-approved drugs to find a therapeutic option for COVID-19. The similarity in SARS-CoV-2 and HIV-1 fusion mechanism to host cells can be a key point for Inhibit SARS-CoV-2 entry into host cells by HIV fusion inhibitors. Accordingly, in this study, an HIV-1 fusion inhibitor called Enfuvirtide (Enf) was selected. The affinity and essential residues involving in the Enf binding to the S2 protein of SARS-CoV-2, HIV-1 gp41 protein and angiotensin-converting enzyme 2 (ACE-2) as a negative control, was evaluated using molecular docking. Eventually, Enf-S2 and Enf-gp41 protein complexes were simulated by molecular dynamics (MD) in terms of binding affinity and stability. Based on the most important criteria such as docking score, cluster size, energy and dissociation constant, the strongest interaction was observed between Enf with the S2 protein. In addition, MD results confirmed that Enf-S2 protein interaction was remarkably stable and caused the S2 protein residues to undergo the fewest fluctuations. In conclusion, it can be stated that Enf can act as a strong SARS-CoV-2 fusion inhibitor and demonstrates the potential to enter the clinical trial phase of COVID-19. Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Enfuvirtide; SARS-CoV-2; in silico drug repurposing; molecular docking; molecular dynamics.
Conflict of interest statement
The authors confirm that this article content has no conflict of interest.
Figures



References
-
- Akya, A., Farasat, A., Ghadiri, K., & Rostamian, M. (2019). Identification of HLA-I restricted epitopes in six vaccine candidates of Leishmania tropica using immunoinformatics and molecular dynamics simulation approaches. Infection, Genetics and Evolution, 75, 103953. 10.1016/j.meegid.2019.103953 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
