High-resolution mapping of the neutralizing and binding specificities of polyclonal sera post-HIV Env trimer vaccination
- PMID: 33438580
- PMCID: PMC7864656
- DOI: 10.7554/eLife.64281
High-resolution mapping of the neutralizing and binding specificities of polyclonal sera post-HIV Env trimer vaccination
Abstract
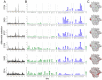
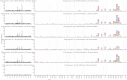
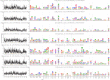
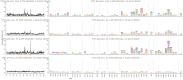
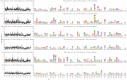
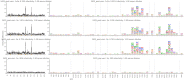
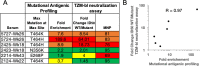
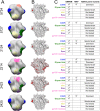
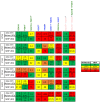
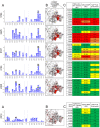
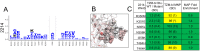
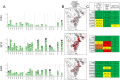
Mapping polyclonal serum responses is critical to rational vaccine design. However, most high-resolution mapping approaches involve isolating and characterizing individual antibodies, which incompletely defines the polyclonal response. Here we use two complementary approaches to directly map the specificities of the neutralizing and binding antibodies of polyclonal anti-HIV-1 sera from rabbits immunized with BG505 Env SOSIP trimers. We used mutational antigenic profiling to determine how all mutations in Env affected viral neutralization and electron microscopy polyclonal epitope mapping (EMPEM) to directly visualize serum Fabs bound to Env trimers. The dominant neutralizing specificities were generally only a subset of the more diverse binding specificities. Additional differences between binding and neutralization reflected antigenicity differences between virus and soluble Env trimer. Furthermore, we refined residue-level epitope specificity directly from sera, revealing subtle differences across sera. Together, mutational antigenic profiling and EMPEM yield a holistic view of the binding and neutralizing specificity of polyclonal sera.
Keywords: Env trimer immunization; HIV; electron microscopy polyclonal epitope mapping; immunology; infectious disease; inflammation; microbiology; mutational antigenic profiling; polyclonal sera; virus.
Plain language summary
Vaccines work by stimulating the immune system to produce proteins called antibodies. These antibodies bind to the virus targeted by the vaccine and block the virus from infecting cells. It has been difficult to develop a vaccine for HIV because frequent mutations allow it to evade antibodies. Understanding exactly how these proteins bind to HIV and how various mutations enable the virus to escape them is crucial to designing a successful HIV vaccine. Over the last decade, scientists have developed new techniques for studying individual antibodies and how they bind to viruses. Now, they are using these insights to design vaccines. Most vaccines result in the production of many antibodies that bind to different parts of the virus, making it harder for a virus to escape. But studying many antibodies with different targets on the virus simultaneously remains challenging. By combining two-cutting edge approaches, Dingens et al. catalogued the many antibodies that rabbits produce in response to an experimental vaccine for HIV. In the experiments, they mapped how two types of rabbit antibodies target the virus: those that could bind to the virus, and those that could both bind and neutralize the virus (i.e., block it from infecting cells). The experiments showed that small differences between the HIV virus and the vaccine explained why some rabbit antibodies created in response to the vaccine could bind but not neutralize the virus. Moreover, the ability to stop HIV from infecting the cells appeared to be reserved to antibodies that could bind to several different locations at the virus. Dingens et al. further documented all the virus mutations that would allow it to evade neutralizing antibodies. The techniques used in the experiments may help scientists identify the best sites on the HIV virus to target with vaccines and to better understand the binding and neutralizing activity of antibodies. The results of the experiments may also help to redesign the experimental HIV vaccine – which is currently being tested in humans – to be even more effective.
© 2021, Dingens et al.
Conflict of interest statement
AD, PP, KM, SH, TK, CC, JM, PK, AW, JB No competing interests declared, JO Reviewing editor, eLife
Figures




References
-
- Alam SM, Aussedat B, Vohra Y, Meyerhoff RR, Cale EM, Walkowicz WE, Radakovich NA, Anasti K, Armand L, Parks R, Sutherland L, Scearce R, Joyce MG, Pancera M, Druz A, Georgiev IS, Von Holle T, Eaton A, Fox C, Reed SG, Louder M, Bailer RT, Morris L, Abdool-Karim SS, Cohen M, Liao HX, Montefiori DC, Park PK, Fernández-Tejada A, Wiehe K, Santra S, Kepler TB, Saunders KO, Sodroski J, Kwong PD, Mascola JR, Bonsignori M, Moody MA, Danishefsky S, Haynes BF. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Science Translational Medicine. 2017;9:eaai7521. doi: 10.1126/scitranslmed.aai7521. - DOI - PMC - PubMed
-
- Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, Lorenzi JCC, Finkin S, Hägglöf T, Hurley A, Millard KG, Weisblum Y, Schmidt F, Hatziioannou T, Bieniasz PD, Caskey M, Robbiani DF, Nussenzweig MC, Bjorkman PJ. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. - DOI - PMC - PubMed
-
- Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, Burton DR, Ward AB, Hangartner L. Electron-Microscopy-Based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity. 2018;49:288–300. doi: 10.1016/j.immuni.2018.07.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

