Molecular mechanisms and physiological functions of mitophagy
- PMID: 33438778
- PMCID: PMC7849173
- DOI: 10.15252/embj.2020104705
Molecular mechanisms and physiological functions of mitophagy
Abstract
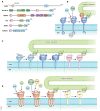
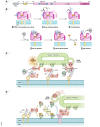
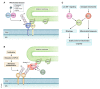
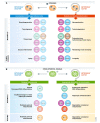
Degradation of mitochondria via a selective form of autophagy, named mitophagy, is a fundamental mechanism conserved from yeast to humans that regulates mitochondrial quality and quantity control. Mitophagy is promoted via specific mitochondrial outer membrane receptors, or ubiquitin molecules conjugated to proteins on the mitochondrial surface leading to the formation of autophagosomes surrounding mitochondria. Mitophagy-mediated elimination of mitochondria plays an important role in many processes including early embryonic development, cell differentiation, inflammation, and apoptosis. Recent advances in analyzing mitophagy in vivo also reveal high rates of steady-state mitochondrial turnover in diverse cell types, highlighting the intracellular housekeeping role of mitophagy. Defects in mitophagy are associated with various pathological conditions such as neurodegeneration, heart failure, cancer, and aging, further underscoring the biological relevance. Here, we review our current molecular understanding of mitophagy, and its physiological implications, and discuss how multiple mitophagy pathways coordinately modulate mitochondrial fitness and populations.
Keywords: autophagy; mitochondria; phosphorylation; quality and quantity control; ubiquitin.
© 2021 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Agnihotri S, Golbourn B, Huang X, Remke M, Younger S, Cairns RA, Chalil A, Smith CA, Krumholtz SL, Mackenzie D et al (2016) PINK1 is a negative regulator of growth and the Warburg effect in glioblastoma. Cancer Res 76: 4708–4719 - PubMed
-
- Al Rawi S, Louvet‐Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V (2011) Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334: 1144–1147 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

