Evidence for S2 flexibility by direct visualization of quantum dot-labeled myosin heads and rods within smooth muscle myosin filaments moving on actin in vitro
- PMID: 33439241
- PMCID: PMC7809879
- DOI: 10.1085/jgp.202012751
Evidence for S2 flexibility by direct visualization of quantum dot-labeled myosin heads and rods within smooth muscle myosin filaments moving on actin in vitro
Abstract
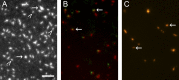
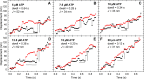
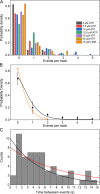
Myosins in muscle assemble into filaments by interactions between the C-terminal light meromyosin (LMM) subdomains of the coiled-coil rod domain. The two head domains are connected to LMM by the subfragment-2 (S2) subdomain of the rod. Our mixed kinetic model predicts that the flexibility and length of S2 that can be pulled away from the filament affects the maximum distance working heads can move a filament unimpeded by actin-attached heads. It also suggests that it should be possible to observe a head remain stationary relative to the filament backbone while bound to actin (dwell), followed immediately by a measurable jump upon detachment to regain the backbone trajectory. We tested these predictions by observing filaments moving along actin at varying ATP using TIRF microscopy. We simultaneously tracked two different color quantum dots (QDs), one attached to a regulatory light chain on the lever arm and the other attached to an LMM in the filament backbone. We identified events (dwells followed by jumps) by comparing the trajectories of the QDs. The average dwell times were consistent with known kinetics of the actomyosin system, and the distribution of the waiting time between observed events was consistent with a Poisson process and the expected ATPase rate. Geometric constraints suggest a maximum of ∼26 nm of S2 can be unzipped from the filament, presumably involving disruption in the coiled-coil S2, a result consistent with observations by others of S2 protruding from the filament in muscle. We propose that sufficient force is available from the working heads in the filament to overcome the stiffness imposed by filament-S2 interactions.
© 2021 Brizendine et al.
Figures





References
-
- Alamo, L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

