Global changes to HepG2 cell metabolism in response to galactose treatment
- PMID: 33439775
- PMCID: PMC8163570
- DOI: 10.1152/ajpcell.00460.2020
Global changes to HepG2 cell metabolism in response to galactose treatment
Abstract
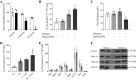
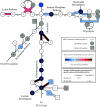
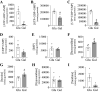
Tumor cell proliferation requires sufficient metabolic flux through the pentose phosphate pathway to meet the demand for biosynthetic precursors and to increase protection against oxidative stress which in turn requires an upregulation of substrate flow through glycolysis. This metabolic poise is often coupled with a shift in ATP production from mitochondrial OXPHOS to substrate-level phosphorylation. Despite major advances that were facilitated by using tumor-derived cell lines in research areas spanning from membrane to cytoskeletal biology, this distorted metabolic profile limits their impact as a model in physiology and toxicology. Substitution of glucose with galactose in the cell culture medium has been demonstrated to shift ATP production from substrate-level phosphorylation to mitochondrial OXPHOS. This increase in oxygen utilization is coupled to a global metabolic reorganization with potential impacts on macromolecule biosynthesis and cellular redox homeostasis, but a comprehensive analysis on the effects of sugar substitution in tumor-derived cells is still missing. To address this gap in knowledge we performed transcriptomic and metabolomic analyses on human hepatocellular carcinoma (HepG2) cells adapted to either glucose or galactose as the aldohexose source. We observed a shift toward oxidative metabolism in all primary metabolic pathways at both transcriptomic and metabolomic levels. We also observed a decrease in nicotinamide dinucleotide (NAD(P)) levels and subcellular NAD+-to-NADH ratios in cells cultured with galactose compared with glucose control cells. Our results suggest that galactose reduces both glycolytic and biosynthetic flux and restores a metabolic poise in HepG2 cells that closely reflects the metabolic state observed in primary hepatocytes.
Keywords: HepG2; NAD; galactose; mitochondria; redox state.
Figures



Similar articles
-
SR4 Uncouples Mitochondrial Oxidative Phosphorylation, Modulates AMP-dependent Kinase (AMPK)-Mammalian Target of Rapamycin (mTOR) Signaling, and Inhibits Proliferation of HepG2 Hepatocarcinoma Cells.J Biol Chem. 2015 Dec 18;290(51):30321-41. doi: 10.1074/jbc.M115.686352. Epub 2015 Nov 3. J Biol Chem. 2015. PMID: 26534958 Free PMC article.
-
Sodium valproate induces mitochondrial respiration dysfunction in HepG2 in vitro cell model.Toxicology. 2015 May 4;331:47-56. doi: 10.1016/j.tox.2015.03.001. Epub 2015 Mar 5. Toxicology. 2015. PMID: 25745980
-
β-Catenin regulates hepatic mitochondrial function and energy balance in mice.Gastroenterology. 2012 Sep;143(3):754-764. doi: 10.1053/j.gastro.2012.05.048. Epub 2012 Jun 7. Gastroenterology. 2012. PMID: 22684045 Free PMC article.
-
Mitochondrial Metabolic Signatures in Hepatocellular Carcinoma.Cells. 2021 Jul 27;10(8):1901. doi: 10.3390/cells10081901. Cells. 2021. PMID: 34440674 Free PMC article. Review.
-
Clear cell hepatocellular carcinoma: origin, metabolic traits and fate of glycogenotic clear and ground glass cells.Hepatobiliary Pancreat Dis Int. 2017 Dec 15;16(6):570-594. doi: 10.1016/S1499-3872(17)60071-7. Hepatobiliary Pancreat Dis Int. 2017. PMID: 29291777 Review.
Cited by
-
Administration of Secretome Derived from Human Mesenchymal Stem Cells Induces Hepatoprotective Effects in Models of Idiosyncratic Drug-Induced Liver Injury Caused by Amiodarone or Tamoxifen.Cells. 2023 Feb 16;12(4):636. doi: 10.3390/cells12040636. Cells. 2023. PMID: 36831304 Free PMC article.
-
A Metabolomic Investigation of Eugenol on Colorectal Cancer Cell Line HT-29 by Modifying the Expression of APC, p53, and KRAS Genes.Evid Based Complement Alternat Med. 2021 Nov 18;2021:1448206. doi: 10.1155/2021/1448206. eCollection 2021. Evid Based Complement Alternat Med. 2021. PMID: 34840582 Free PMC article.
-
Multifaceted roles of aerobic glycolysis and oxidative phosphorylation in hepatocellular carcinoma.PeerJ. 2023 Feb 1;11:e14797. doi: 10.7717/peerj.14797. eCollection 2023. PeerJ. 2023. PMID: 36748090 Free PMC article. Review.
-
Exploring Dynamic Metabolome of the HepG2 Cell Line: Rise and Fall.Cells. 2022 Nov 10;11(22):3548. doi: 10.3390/cells11223548. Cells. 2022. PMID: 36428976 Free PMC article.
-
Metabolic Priming as a Tool in Redox and Mitochondrial Theragnostics.Antioxidants (Basel). 2023 May 10;12(5):1072. doi: 10.3390/antiox12051072. Antioxidants (Basel). 2023. PMID: 37237939 Free PMC article. Review.
References
-
- Warburg O. On respiratory impairment in cancer cells. Science 124: 269–270, 1956. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

