Analysis of the Immune Responses in the Ileum of Gnotobiotic Pigs Infected with the Recombinant GII.p12_GII.3 Human Norovirus by mRNA Sequencing
- PMID: 33440894
- PMCID: PMC7826840
- DOI: 10.3390/v13010092
Analysis of the Immune Responses in the Ileum of Gnotobiotic Pigs Infected with the Recombinant GII.p12_GII.3 Human Norovirus by mRNA Sequencing
Abstract
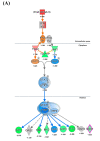
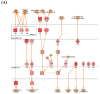
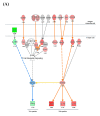
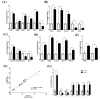
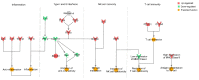
Norovirus genogroup II (NoV GII) induces acute gastrointestinal food-borne illness in humans. Because gnotobiotic pigs can be infected with human norovirus (HuNoV) GII, they are frequently used to analyze the associated pathogenic mechanisms and immune responses, which remain poorly understood. Recently, mRNA sequencing analysis (RNA-Seq) has been used to identify cellular responses to viruses. In this study, we investigated the host immune response and possible mechanisms involved in virus evasion in the ileum of gnotobiotic pigs infected with HuNoV by RNA-Seq. HuNoV was detected in the feces, blood, and tissues of the jejunum, ileum, colon, mesenteric lymph node, and spleen of pigs infected with HuNoV. In analysis of mRNA sequencing, expression of anti-viral protein genes such as OAS1, MX1, and MX2 were largely decreased, whereas type I IFN was increased in pigs infected with HuNoV. In addition, expression of TNF and associated anti-inflammatory cytokine genes such as IL10 was increased in HuNoV-infected pigs. Expression of genes related to natural killer (NK) cell cytotoxicity and CD8+ T cell exhaustion was increased, whereas that of MHC class I genes was decreased. Expression profiles of selected genes were further confirmed by qRT-PCR and Western blot. These results suggest that infection with HuNoV induces NK cell-mediated cytotoxicity but suppresses type I IFN- and CD8+ T cell-mediated antiviral responses.
Keywords: gnotobiotic pig; high-throughput mRNA sequencing; human norovirus; immune response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

