The 3- O-sulfation of heparan sulfate modulates protein binding and lyase degradation
- PMID: 33441484
- PMCID: PMC7826381
- DOI: 10.1073/pnas.2012935118
The 3- O-sulfation of heparan sulfate modulates protein binding and lyase degradation
Abstract
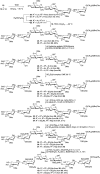
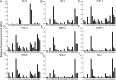
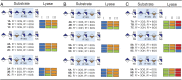
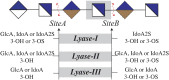
Humans express seven heparan sulfate (HS) 3-O-sulfotransferases that differ in substrate specificity and tissue expression. Although genetic studies have indicated that 3-O-sulfated HS modulates many biological processes, ligand requirements for proteins engaging with HS modified by 3-O-sulfate (3-OS) have been difficult to determine. In particular, the context in which the 3-OS group needs to be presented for binding is largely unknown. We describe herein a modular synthetic approach that can provide structurally diverse HS oligosaccharides with and without 3-OS. The methodology was employed to prepare 27 hexasaccharides that were printed as a glycan microarray to examine ligand requirements of a wide range of HS-binding proteins. The binding selectivity of antithrombin-III (AT-III) compared well with anti-Factor Xa activity supporting robustness of the array technology. Many of the other examined HS-binding proteins required an IdoA2S-GlcNS3S6S sequon for binding but exhibited variable dependence for the 2-OS and 6-OS moieties, and a GlcA or IdoA2S residue neighboring the central GlcNS3S. The HS oligosaccharides were also examined as inhibitors of cell entry by herpes simplex virus type 1, which, surprisingly, showed a lack of dependence of 3-OS, indicating that, instead of glycoprotein D (gD), they competitively bind to gB and gC. The compounds were also used to examine substrate specificities of heparin lyases, which are enzymes used for depolymerization of HS/heparin for sequence determination and production of therapeutic heparins. It was found that cleavage by lyase II is influenced by 3-OS, while digestion by lyase I is only affected by 2-OS. Lyase III exhibited sensitivity to both 3-OS and 2-OS.
Keywords: 3-O-sulfation; anti-Factor Xa; glycan microarray; heparin lyases; herpes simplex virus 1.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Esko J. D., Selleck S. B., Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 (2002). - PubMed
-
- Bishop J. R., Schuksz M., Esko J. D., Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 (2007). - PubMed
-
- Sasisekharan R., Venkataraman G., Heparin and heparan sulfate: Biosynthesis, structure and function. Curr. Opin. Chem. Biol. 4, 626–631 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

