Color-tuning of natural variants of heliorhodopsin
- PMID: 33441566
- PMCID: PMC7807009
- DOI: 10.1038/s41598-020-72125-0
Color-tuning of natural variants of heliorhodopsin
Abstract
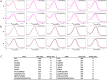
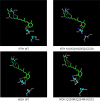
Microbial rhodopsins are distributed through many microorganisms. Heliorhodopsins are newly discovered but have an unclear function. They have seven transmembrane helices similar to type-I and type-II rhodopsins, but they are different in that the N-terminal region of heliorhodopsin is cytoplasmic. We chose 13 representative heliorhodopsins from various microorganisms, expressed and purified with an N-terminal His tag, and measured the absorption spectra. The 13 natural variants had an absorption maximum (λmax) in the range 530-556 nm similar to proteorhodopsin (λmax = 490-525 nm). We selected several candidate residues that influence rhodopsin color-tuning based on sequence alignment and constructed mutants via site-directed mutagenesis to confirm the spectral changes. We found two important residues located near retinal chromophore that influence λmax. We also predict the 3D structure via homology-modeling of Thermoplasmatales heliorhodopsin. The results indicate that the color-tuning mechanism of type-I rhodopsin can be applied to understand the color-tuning of heliorhodopsin.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

