Structural and functional analysis of the Francisella lysine decarboxylase as a key actor in oxidative stress resistance
- PMID: 33441661
- PMCID: PMC7806604
- DOI: 10.1038/s41598-020-79611-5
Structural and functional analysis of the Francisella lysine decarboxylase as a key actor in oxidative stress resistance
Abstract
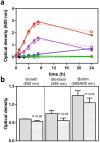
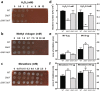
Francisella tularensis is one of the most virulent pathogenic bacteria causing the acute human respiratory disease tularemia. While the mechanisms underlying F. tularensis pathogenesis are largely unknown, previous studies have shown that a F. novicida transposon mutant with insertions in a gene coding for a putative lysine decarboxylase was attenuated in mouse spleen, suggesting a possible role of its protein product as a virulence factor. Therefore, we set out to structurally and functionally characterize the F. novicida lysine decarboxylase, which we termed LdcF. Here, we investigate the genetic environment of ldcF as well as its evolutionary relationships with other basic AAT-fold amino acid decarboxylase superfamily members, known as key actors in bacterial adaptative stress response and polyamine biosynthesis. We determine the crystal structure of LdcF and compare it with the most thoroughly studied lysine decarboxylase, E. coli LdcI. We analyze the influence of ldcF deletion on bacterial growth under different stress conditions in dedicated growth media, as well as in infected macrophages, and demonstrate its involvement in oxidative stress resistance. Finally, our mass spectrometry-based quantitative proteomic analysis enables identification of 80 proteins with expression levels significantly affected by ldcF deletion, including several DNA repair proteins potentially involved in the diminished capacity of the F. novicida mutant to deal with oxidative stress. Taken together, we uncover an important role of LdcF in F. novicida survival in host cells through participation in oxidative stress response, thereby singling out this previously uncharacterized protein as a potential drug target.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
An AraC/XylS Family Transcriptional Regulator Modulates the Oxidative Stress Response of Francisella tularensis.J Bacteriol. 2021 Nov 5;203(23):e0018521. doi: 10.1128/JB.00185-21. Epub 2021 Sep 20. J Bacteriol. 2021. PMID: 34543107 Free PMC article.
-
Elucidation of a mechanism of oxidative stress regulation in Francisella tularensis live vaccine strain.Mol Microbiol. 2016 Sep;101(5):856-78. doi: 10.1111/mmi.13426. Epub 2016 Jun 16. Mol Microbiol. 2016. PMID: 27205902 Free PMC article.
-
Characterization of a Unique Outer Membrane Protein Required for Oxidative Stress Resistance and Virulence of Francisella tularensis.J Bacteriol. 2018 Mar 26;200(8):e00693-17. doi: 10.1128/JB.00693-17. Print 2018 Apr 15. J Bacteriol. 2018. PMID: 29378894 Free PMC article.
-
Comparative review of Francisella tularensis and Francisella novicida.Front Cell Infect Microbiol. 2014 Mar 13;4:35. doi: 10.3389/fcimb.2014.00035. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24660164 Free PMC article. Review.
-
Proteins involved in Francisella tularensis survival and replication inside macrophages.Future Microbiol. 2012 Nov;7(11):1255-68. doi: 10.2217/fmb.12.103. Future Microbiol. 2012. PMID: 23075445 Review.
Cited by
-
Filamentation-driven peripheral clustering of the inducible lysine decarboxylase is crucial for E. coli acid stress response.Commun Biol. 2025 Aug 6;8(1):1168. doi: 10.1038/s42003-025-08616-5. Commun Biol. 2025. PMID: 40770054 Free PMC article.
-
The Biosynthetic Pathway of Ubiquinone Contributes to Pathogenicity of Francisella novicida.J Bacteriol. 2021 Nov 5;203(23):e0040021. doi: 10.1128/JB.00400-21. Epub 2021 Sep 20. J Bacteriol. 2021. PMID: 34543102 Free PMC article.
-
Pathogenicity and virulence of Francisella tularensis.Virulence. 2023 Dec;14(1):2274638. doi: 10.1080/21505594.2023.2274638. Epub 2023 Nov 8. Virulence. 2023. PMID: 37941380 Free PMC article. Review.
-
Microbial adaptive evolution.J Ind Microbiol Biotechnol. 2022 Apr 14;49(2):kuab076. doi: 10.1093/jimb/kuab076. J Ind Microbiol Biotechnol. 2022. PMID: 34673973 Free PMC article. Review.
-
α-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?ACS Catal. 2025 May 2;15(10):8204-8218. doi: 10.1021/acscatal.5c00326. eCollection 2025 May 16. ACS Catal. 2025. PMID: 40401103
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

