Simultaneous lesion and brain segmentation in multiple sclerosis using deep neural networks
- PMID: 33441684
- PMCID: PMC7806997
- DOI: 10.1038/s41598-020-79925-4
Simultaneous lesion and brain segmentation in multiple sclerosis using deep neural networks
Abstract
Segmentation of white matter lesions and deep grey matter structures is an important task in the quantification of magnetic resonance imaging in multiple sclerosis. In this paper we explore segmentation solutions based on convolutional neural networks (CNNs) for providing fast, reliable segmentations of lesions and grey-matter structures in multi-modal MR imaging, and the performance of these methods when applied to out-of-centre data. We trained two state-of-the-art fully convolutional CNN architectures on the 2016 MSSEG training dataset, which was annotated by seven independent human raters: a reference implementation of a 3D Unet, and a more recently proposed 3D-to-2D architecture (DeepSCAN). We then retrained those methods on a larger dataset from a single centre, with and without labels for other brain structures. We quantified changes in performance owing to dataset shift, and changes in performance by adding the additional brain-structure labels. We also compared performance with freely available reference methods. Both fully-convolutional CNN methods substantially outperform other approaches in the literature when trained and evaluated in cross-validation on the MSSEG dataset, showing agreement with human raters in the range of human inter-rater variability. Both architectures showed drops in performance when trained on single-centre data and tested on the MSSEG dataset. When trained with the addition of weak anatomical labels derived from Freesurfer, the performance of the 3D Unet degraded, while the performance of the DeepSCAN net improved. Overall, the DeepSCAN network predicting both lesion and anatomical labels was the best-performing network examined.
Conflict of interest statement
Richard McKinley, Rik Wepfer, Fabian Aschwanden, Lorenz Grunder, Raphaela Muri, Christian Rummel, Rajeev Verma, Christian Weisstanner, Mauricio Reyes, Franca Wagner and Roland Wiest have no interests to declare. Anke Salmen has recieved speaker honoraria and/or travel compensation for activities with Almirall Hermal GmbH, Biogen, Merck, Novartis, Roche, and Sanofi Genzyme, none related to this work. Andrew Chan received honoraria for board and speaker honoraria from Actelion, Bayer, Biogen, Celgene, Merck, Novartis, Sanofi-Genzyme, Roche, Teva, all for hospital/university research funds. He has recieved reserach funds from Research funds: UCB, Biogen, Sanofi-Genzyme, and is an editor for European Journal of Neurology, and Clin Transl Neurosci.
Figures
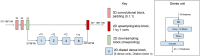

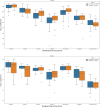

References
-
- McFarland, H. F. et al. Using gadolinium-enhanced magnetic resonance imaging lesions to monitor disease activity in multiple sclerosis. Ann. Neurol.32, 758–766, 10.1002/ana.410320609 (1992). - PubMed
-
- Wattjes MP, et al. Evidence-based guidelines: Magnims consensus guidelines on the use of mri in multiple sclerosis—establishing disease prognosis and monitoring patients. Nat. Rev. Neurol. 2015;11:597–606. - PubMed
-
- McKinley R, et al. et al. Nabla-net: A deep dag-like convolutional architecture for biomedical image segmentation. In: Crimi A, et al.et al., editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2016. pp. 119–128.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

