Histone acetyltransferase PfGCN5 regulates stress responsive and artemisinin resistance related genes in Plasmodium falciparum
- PMID: 33441725
- PMCID: PMC7806804
- DOI: 10.1038/s41598-020-79539-w
Histone acetyltransferase PfGCN5 regulates stress responsive and artemisinin resistance related genes in Plasmodium falciparum
Abstract
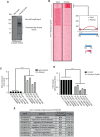
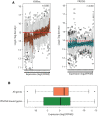
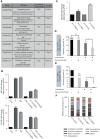
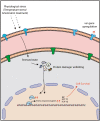
Plasmodium falciparum has evolved resistance to almost all front-line drugs including artemisinin, which threatens malaria control and elimination strategies. Oxidative stress and protein damage responses have emerged as key players in the generation of artemisinin resistance. In this study, we show that PfGCN5, a histone acetyltransferase, binds to the stress-responsive genes in a poised state and regulates their expression under stress conditions. Furthermore, we show that upon artemisinin exposure, genome-wide binding sites for PfGCN5 are increased and it is directly associated with the genes implicated in artemisinin resistance generation like BiP and TRiC chaperone. Interestingly, expression of genes bound by PfGCN5 was found to be upregulated during stress conditions. Moreover, inhibition of PfGCN5 in artemisinin-resistant parasites increases the sensitivity of the parasites to artemisinin treatment indicating its role in drug resistance generation. Together, these findings elucidate the role of PfGCN5 as a global chromatin regulator of stress-responses with a potential role in modulating artemisinin drug resistance and identify PfGCN5 as an important target against artemisinin-resistant parasites.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Plasmodium falciparum GCN5 plays a key role in regulating artemisinin resistance-related stress responses.Antimicrob Agents Chemother. 2023 Oct 18;67(10):e0057723. doi: 10.1128/aac.00577-23. Epub 2023 Sep 13. Antimicrob Agents Chemother. 2023. PMID: 37702516 Free PMC article.
-
Role of PfGCN5 in nutrient sensing and transcriptional regulation in Plasmodium falciparum.J Biosci. 2020;45:11. J Biosci. 2020. PMID: 31965989
-
Plasmodium berghei K13 Mutations Mediate In Vivo Artemisinin Resistance That Is Reversed by Proteasome Inhibition.mBio. 2020 Nov 10;11(6):e02312-20. doi: 10.1128/mBio.02312-20. mBio. 2020. PMID: 33173001 Free PMC article.
-
A brief history of artemisinin: Modes of action and mechanisms of resistance.Chin J Nat Med. 2019 May 20;17(5):331-336. doi: 10.1016/S1875-5364(19)30038-X. Chin J Nat Med. 2019. PMID: 31171267 Review.
-
Plasmodium falciparum kelch 13: a potential molecular marker for tackling artemisinin-resistant malaria parasites.Expert Rev Anti Infect Ther. 2016;14(1):125-35. doi: 10.1586/14787210.2016.1106938. Epub 2015 Nov 4. Expert Rev Anti Infect Ther. 2016. PMID: 26535806 Review.
Cited by
-
PfGCN5 is essential for Plasmodium falciparum survival and transmission and regulates Pf H2B.Z acetylation and chromatin structure.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf218. doi: 10.1093/nar/gkaf218. Nucleic Acids Res. 2025. PMID: 40156869 Free PMC article.
-
Epigenetic regulation as a therapeutic target in the malaria parasite Plasmodium falciparum.Malar J. 2024 Feb 12;23(1):44. doi: 10.1186/s12936-024-04855-9. Malar J. 2024. PMID: 38347549 Free PMC article. Review.
-
Pervasive sequence-level variation in the transcriptome of Plasmodium falciparum.NAR Genom Bioinform. 2022 May 17;4(2):lqac036. doi: 10.1093/nargab/lqac036. eCollection 2022 Jun. NAR Genom Bioinform. 2022. PMID: 35591889 Free PMC article.
-
The Putative Bromodomain Protein PfBDP7 of the Human Malaria Parasite Plasmodium Falciparum Cooperates With PfBDP1 in the Silencing of Variant Surface Antigen Expression.Front Cell Dev Biol. 2022 Apr 12;10:816558. doi: 10.3389/fcell.2022.816558. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35493110 Free PMC article.
-
GCN5 mediates DNA-PKcs crotonylation for DNA double-strand break repair and determining cancer radiosensitivity.Br J Cancer. 2024 Jun;130(10):1621-1634. doi: 10.1038/s41416-024-02636-4. Epub 2024 Apr 4. Br J Cancer. 2024. PMID: 38575732 Free PMC article.
References
-
- Organization, W. H. World malaria report 2019. (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

