Tethering-induced destabilization and ATP-binding for tandem RRM domains of ALS-causing TDP-43 and hnRNPA1
- PMID: 33441818
- PMCID: PMC7806782
- DOI: 10.1038/s41598-020-80524-6
Tethering-induced destabilization and ATP-binding for tandem RRM domains of ALS-causing TDP-43 and hnRNPA1
Abstract
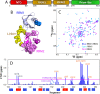
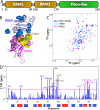
TDP-43 and hnRNPA1 contain tandemly-tethered RNA-recognition-motif (RRM) domains, which not only functionally bind an array of nucleic acids, but also participate in aggregation/fibrillation, a pathological hallmark of various human diseases including amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), alzheimer's disease (AD) and Multisystem proteinopathy (MSP). Here, by DSF, NMR and MD simulations we systematically characterized stability, ATP-binding and conformational dynamics of TDP-43 and hnRNPA1 RRM domains in both tethered and isolated forms. The results reveal three key findings: (1) upon tethering TDP-43 RRM domains become dramatically coupled and destabilized with Tm reduced to only 49 °C. (2) ATP specifically binds TDP-43 and hnRNPA1 RRM domains, in which ATP occupies the similar pockets within the conserved nucleic-acid-binding surfaces, with the affinity slightly higher to the tethered than isolated forms. (3) MD simulations indicate that the tethered RRM domains of TDP-43 and hnRNPA1 have higher conformational dynamics than the isolated forms. Two RRM domains become coupled as shown by NMR characterization and analysis of inter-domain correlation motions. The study explains the long-standing puzzle that the tethered TDP-43 RRM1-RRM2 is particularly prone to aggregation/fibrillation, and underscores the general role of ATP in inhibiting aggregation/fibrillation of RRM-containing proteins. The results also rationalize the observation that the risk of aggregation-causing diseases increases with aging.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

