Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq
- PMID: 33441952
- PMCID: PMC7806779
- DOI: 10.1038/s41598-020-80881-2
Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq
Abstract
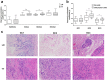
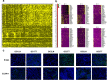
Gastric cancer (GC) is a leading cause of cancer-induced mortality, with poor prognosis with metastasis. The mechanism of gastric carcinoma lymph node metastasis remains unknown due to traditional bulk-leveled approaches masking the roles of subpopulations. To answer questions concerning metastasis from the gastric carcinoma intratumoural perspective, we performed single-cell level analysis on three gastric cancer patients with primary cancer and paired metastatic lymph node cancer tissues using single-cell RNA-seq (scRNA-seq). The results showed distinct carcinoma profiles from each patient, and diverse microenvironmental subsets were shared across different patients. Clustering data showed significant intratumoural heterogeneity. The results also revealed a subgroup of cells bridging the metastatic group and primary group, implying the transition state of cancer during the metastatic process. In the present study, we obtained a more comprehensive picture of gastric cancer lymph node metastasis, and we discovered some GC lymph node metastasis marker genes (ERBB2, CLDN11 and CDK12), as well as potential gastric cancer evolution-driving genes (FOS and JUN), which provide a basis for the treatment of GC.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

